Introduction to data handling and visualisation in R
This module provides an introduction to R and RStudio. It includes hands on teaching for how to handle data in R using key packages such as tidyverse sf, sp, rgdal and raster.
In this module we will cover what R is, how to install it on your machine and how to interact with the RStudio interface. We will look at how to manage your projects in R then introduce basic data management and processing. This will then be expanded on and you will learn about data manipulation and key functions used to clean and investigate your data. We will introduce the ggplot package and explore how this can be used to produce high quality data visualisations. Finally we will cover how to save your data.
The teaching is followed by a practical session implementing the acquired skills to clean and process replica DHIS2 routine malaria surveillance data.
Introduction to R and Rstudio
R is a popular programming language for data science. It is a powerful language which can be used for basic data cleaning and analysis, building complex statistical models, creating publication quality visualisations, as well building interactive tools and websites. R is an open source software with numerous people contributing packages to improve it”s functionality.
We will be using R to run all of the code in this module. To improve ease of use we will run this through the RStudio interface. RStudio is an integrated development environment (IDE) from which we can run R. This is more user friendly and has large functionality for organising projects and building reproducible code.
Installing R and RStudio
Firstly to install R go to the R-project webpage: https://www.r-project.org/. Select a CRAN mirror in a location close to you from which to download R. Follow the prompts to download R for your processing system.
Rstudio can then be downloaded from the webpage: https://www.rstudio.com/products/rstudio/download. Select the free desktop version to proceed.
Overview of RStudio
Once installed, open up RStudio. You will see the console on your left, this is where you can type code and run R commands. There are two windows on your right, the top one shows you what is in the global R environment, and the bottom one is where you can view files, plots, packages and help pages.
There are two ways we can run R code in RStudio, directly from the console, or from an R script. We will start by looking at the console where we can write basic code to be run line by line.
Project management with Rstudio
In RStudio you can use “projects” to manage your files and code. These help by organising everything into a working directory and allowing you to easily keep your files, scripts and outputs for a single project in one place.
You can either create a new project from scratch in RStudio, create a project from an existing folder containing R code or by cloning a version controlled (e.g. Git) repository.
We will look at how to start a new project in RStudio. To do this we go to File > New Project. Select the option to create the project in a new directory and give the directory a name and create the project.
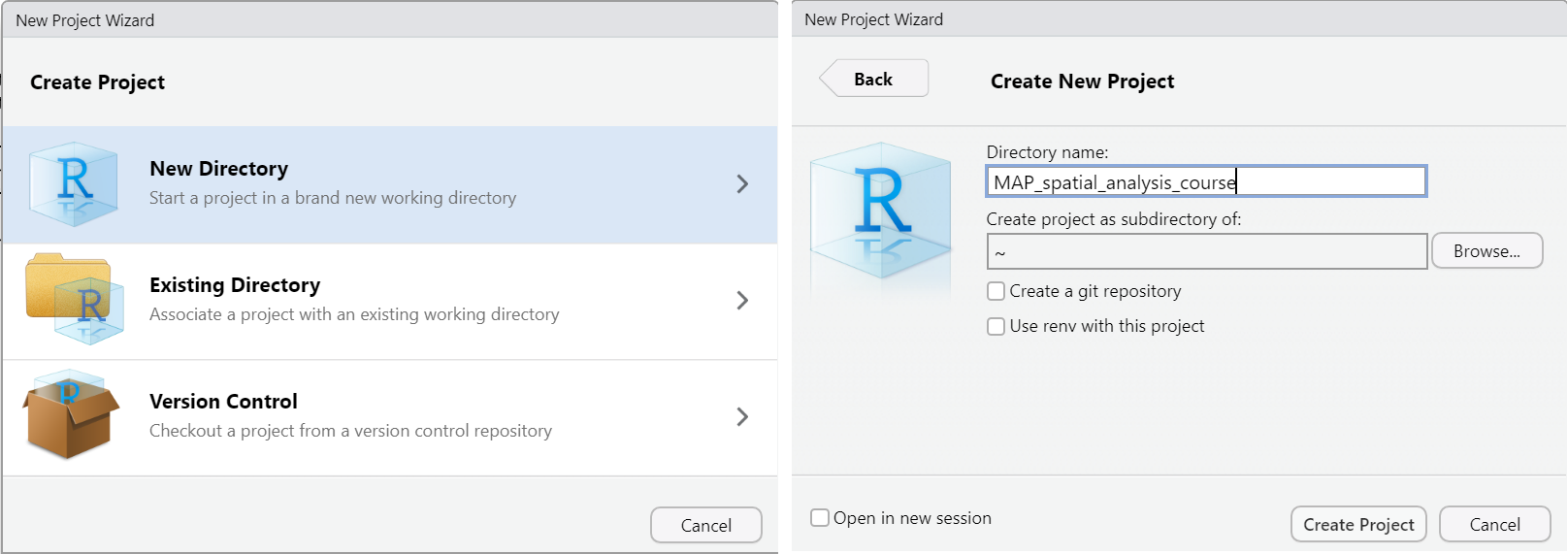
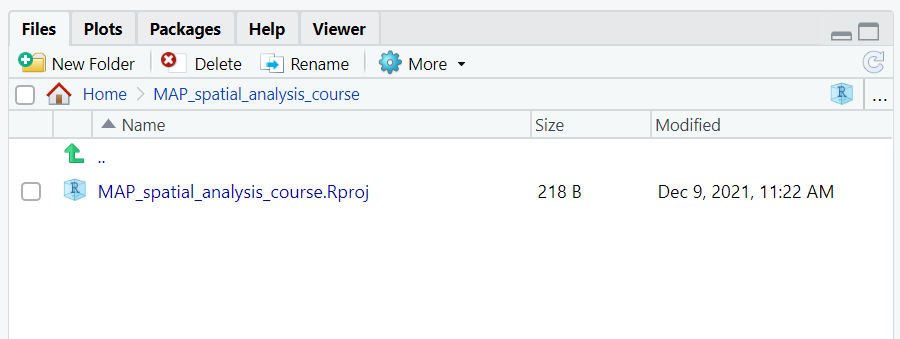
You will then see the project appear in the bottom right window. You can now navigate your project from this window. You can see the files you have saved as part of the project, create new folders, delete, move or rename files.
In this tutorial we will be working with the R project “NAME OF
PROJECT” which can be downloaded from “WEBLINK TO DATA”.
Data types and structures
We will now introduce you to some of the basic data types in R. There are five main types of data in R, these are:
- Characters : “a”, “bananas”
- Numeric: 1 , 2.2321 , 1999
- Integer: 2L, 5L (L tells R to store as integer)
- Logical: True, False
- Complex: 1+4i (I is the imaginary part)
- Factors (categorical data with labels)
The major data structures in R are:
- Vectors
- Matrices
- Arrays
- Data frames
- Lists
Vectors are series of values, either numbers, characters or logical values (TRUE or FALSE).
Matrices are 2-dimensional array which can hold numeric, character or logical data types. All of the data in a matrix has to be of the same type and there is a fixed number of rows and columns.
A array is of 2 or more dimensions and as with a matrix can hold numeric, character or logical data types, with all data being of the same type
Data frames are a selection of equal length vectors grouped together, where the vectors can be of different data types. These are the most common format used to analyse datasets in and what we will primarily be working with in this session.
Lists are essentially a list of other items, these can be a mixture of different data types, for example you can create a list of data frames, matrices and vectors. It is useful way of organising and storing data.
Basic expressions
In the console we can write expressions and execute them by pressing enter. The most basic code we can write is simple calculations; when we press enter the calculation will be processed and a value returned.
5 + 5## [1] 10You can create variables and assign values to them, for example, we
want to create a variable for the number of people tested for malaria.
We want the variable to be called “tested” and for it to have the value
of 120. We assign this using the <- sign. This will now
appear in your global environment window on the top right. We can use
the shortcut “alt and -” to input the
<- sign.
tested <- 120You can then refer to the variable in the code you write and it will work as the value assigned to it. It is then possible to do calculations based on this variable.
tested * 2## [1] 240tested / 10## [1] 12Creating another variable will allow us to perform calculations between those variables. These variables can then be updated and the same code rerun to produce updated results. For example, we received information telling us that of the 120 people tested for malaria, 80 were confirmed positive. We can assign this value to the variable “positive” and calculate the proportion of those tested who were confirmed positive.
positive <- 80
positive / tested ## Proportion of people tested confirmed positive## [1] 0.6666667If we then received updated information that 160 people were tested, not 120, we can update the value for tested and recalculate the proportion of positive cases.
tested <- 160
positive / tested ## [1] 0.5We can store the output value as a new variable and give it a name. This can then be called on from the environment.
proportion_positive <- positive / tested
proportion_positive## [1] 0.5R isn’t restricted to numeric variables, we can also create variables of character strings in the same way.
country <- "Burkina Faso"
country## [1] "Burkina Faso"Vectors
In R we commonly work with vectors, these are sequences of values.
Vectors can be numeric or character strings and are created using the
c(), concatenate, function. Once we have created the vector
we can access different values of the vector by using square brackets.
[2] will return the second value in the vector.
tested <- c(56, 102, 65, 43, 243)
tested[2]## [1] 102countries <- c("Burkina Faso", "Nigeria", "Tanzania", "Uganda")
countries[2]## [1] "Nigeria"Basic functions
Next we can run some functions in R. There are a range of base functions in R, and additional ones made available by installing “packages” (more details on this later). Functions often take arguments in brackets (). Some of the base functions in R allow us to easily compute common summary statistics on vectors. Here are some examples.
sum(tested)## [1] 509mean(tested)## [1] 101.8max(tested)## [1] 243Some functions will take additional arguments. Take the
round() function. This takes two arguments, the first is
the number you wish to round, the second is the number of digits to
round to.
round(3.14159, 2)## [1] 3.14Scripts
In most situations you will be wanting to write multiple lines of
reproducible code. To do this you can open an R script by going to File
> New File > R Script (or the shortcut Ctrl+Shift+N).
This opens a text editor in the top left window where we can write our
code. You can then run the whole script by clicking the “Run” button.
Alternatively, you can run sections by highlighting the lines to execute
and either clicking “Run” or Ctrl+Enter. You can also run
the script line by line, by typing Ctr+Enter to run the
line your cursor is currently on. Scripts can be saved, shared with
others, and built on or rerun at later dates. From this point on we will
work in R scripts.
Installing packages
R has a range of base functions available to be used, these include
things such as mean(), sqrt() and
round(). For more advanced data management, analysis and
visualisations there are additional packages you can download which
contain a wide range of functions.
Many packages are available on CRAN. CRAN is “The Comprehensive R
Archive Network” and it stores up-to-date versions of code and
documentation for R. Packages from CRAN can be installed directly from
RStudio. For this module we will primarily be using the
tidyverse group of packages. First we need to install the
packages, then we need to load it into R to be able to use the
functions. At the top of most R scripts you will find code for the
packages being used in that script.
install.packages("tidyverse")
library(tidyverse)You will only need to install the package onto your machine once, from then on for each new session of R you will need to load the packages required, but not to install them again.
Help available in R studio
There is a range of help available in RStudio. The most basic and
informative of these is the help() function or
? operator. These provide access to the R help pages and
documentation for the packages and functions. For example, if we look at
the help page for the round(), which is a base R function,
we can see a description of the function and the format the code needs
to take.
?roundWe can also access help pages for packages. This will show all of the help pages available for that package
help(package = tidyverse)Alternatively many packages have vignettes. These are documents detailing the facilities in a package. They can be accessed from the help page of the project, as above, or called on directly.
vignette("pivot")Finally, the internet provides a huge amount of help for coding in R. Search engines such as Google will provide multiple results for the problems you face (if you are struggling someone will have had the same problem before and asked for help!). Sites such as stackoverflow contain large numbers of questions and answers from the R users community.
Importing and exploring data in R
Importing data
R has the functionality to import various types of data. One of the
most common file types we work with are .csv files, but we
can also import .dta files and .xlsx files
among others.
We can import .csv files into RStudio using the base function
read.csv(). In this function we insert the file path to the
data we wish to import. Note that as we have set up a project, this is
our base directory so file paths stem from where the project is saved.
We can assign the imported dataset to an object, here called
“routine_data”, using the <- operator to store it in our
environment.
For this module we are using a dataset called “routine_data”. This is an fake dataset of malaria routine case reporting and contains variables for the numbers of malaria tests performed and the numbers of confirmed cases. Data are reported by health facility for children under and over the age of 5 seperately.
routine_data <- read.csv("data/routine_data.csv") Other data types can easily be read into R using different packages
available. Stata files (.dta) can be read in using the
read_dta() function from the Haven package,
and excel files (.xls or .xlsx) using the read_excel()
function from the readxl package. Both of these packages
are part of the tidyverse we installed earlier so they do
not need to be installed, just loaded.
## Examples, not run
library(haven)
dta_file <- read_dta("filename.dta")
library(readxl)
excel_file <- read_excel("filename.xlsx")Whilst read.csv() is the most common way to read data
into R, there is the alternative read_csv() function (note
the underscore). This is part of the tidyverse group of
packages and is often a better option for reading in CSV files. This
function is quicker, it reads the data in as a tibble instead of a data
frame and allows for non standard variable namesm amongst other
benefits.
routine_data <- read_csv("data/routine_data.csv") ## Rows: 1200 Columns: 17
## ── Column specification ────────────────────────────────────────────────────────
## Delimiter: ","
## chr (3): adm1, adm2, month
## dbl (14): hf, year, test_u5, test_rdt_u5, test_mic_u5, conf_u5, conf_rdt_u5,...
##
## ℹ Use `spec()` to retrieve the full column specification for this data.
## ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.There are various options we can use when importing data, such as
whether to include headers, what character to use for decimal points,
what to import as missing values. To explore these options you can look
at the help pages e.g. ?read_csv.
Data frames
We will now look at the csv we just imported. You will now see the
“routine_data” object in your environment that we created and assigned
this dataset too. You can see what the data looks like either by
clicking on it in the global environment window or by typing the command
View(routine_data) which opens up a window displaying the
data. Alternatively, we may just want to look at a few rows. We can do
this by using the head() function, which shows us the first
n rows of data
head(routine_data, 5)## # A tibble: 5 × 17
## adm1 adm2 hf month year test_u5 test_rdt_u5 test_mic_u5 conf_u5
## <chr> <chr> <dbl> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 West Bamakiary 6 Jan 2018 289 279 127 204
## 2 West Bamakiary 6 Feb 2018 178 175 87 92
## 3 West Bamakiary 6 Mar 2018 41 40 13 36
## 4 West Bamakiary 6 Apr 2018 NA 129 49 69
## 5 West Bamakiary 6 May 2018 95 93 28 64
## # … with 8 more variables: conf_rdt_u5 <dbl>, conf_mic_u5 <dbl>,
## # test_ov5 <dbl>, test_rdt_ov5 <dbl>, test_mic_ov5 <dbl>, conf_ov5 <dbl>,
## # conf_rdt_ov5 <dbl>, conf_mic_ov5 <dbl>To understand the structure of the data we can use the
str() command. This shows us that “routine_data” is a
data.frame with 15 observations of 6 variables. The variables “country”
and “method” are character strings, whilst “year”, “tested” and
“confirmed” are integers and “mean_age” is numeric.
str(routine_data)## spec_tbl_df [1,200 × 17] (S3: spec_tbl_df/tbl_df/tbl/data.frame)
## $ adm1 : chr [1:1200] "West" "West" "West" "West" ...
## $ adm2 : chr [1:1200] "Bamakiary" "Bamakiary" "Bamakiary" "Bamakiary" ...
## $ hf : num [1:1200] 6 6 6 6 6 6 6 6 6 6 ...
## $ month : chr [1:1200] "Jan" "Feb" "Mar" "Apr" ...
## $ year : num [1:1200] 2018 2018 2018 2018 2018 ...
## $ test_u5 : num [1:1200] 289 178 41 NA 95 108 121 299 323 526 ...
## $ test_rdt_u5 : num [1:1200] 279 175 40 129 93 105 118 293 317 514 ...
## $ test_mic_u5 : num [1:1200] 127 87 13 49 28 38 46 73 73 76 ...
## $ conf_u5 : num [1:1200] 204 92 36 69 64 42 93 175 174 259 ...
## $ conf_rdt_u5 : num [1:1200] 201 90 35 68 62 41 87 171 171 252 ...
## $ conf_mic_u5 : num [1:1200] 9 4 2 3 4 2 6 6 6 12 ...
## $ test_ov5 : num [1:1200] 317 193 45 137 101 115 134 352 394 684 ...
## $ test_rdt_ov5: num [1:1200] 137 73 25 66 57 58 66 229 265 540 ...
## $ test_mic_ov5: num [1:1200] 63 36 8 25 18 22 25 56 62 80 ...
## $ conf_ov5 : num [1:1200] 272 104 47 79 83 49 108 335 391 554 ...
## $ conf_rdt_ov5: num [1:1200] 255 97 44 74 77 46 101 321 374 525 ...
## $ conf_mic_ov5: num [1:1200] 11 5 2 3 4 2 7 12 14 24 ...
## - attr(*, "spec")=
## .. cols(
## .. adm1 = col_character(),
## .. adm2 = col_character(),
## .. hf = col_double(),
## .. month = col_character(),
## .. year = col_double(),
## .. test_u5 = col_double(),
## .. test_rdt_u5 = col_double(),
## .. test_mic_u5 = col_double(),
## .. conf_u5 = col_double(),
## .. conf_rdt_u5 = col_double(),
## .. conf_mic_u5 = col_double(),
## .. test_ov5 = col_double(),
## .. test_rdt_ov5 = col_double(),
## .. test_mic_ov5 = col_double(),
## .. conf_ov5 = col_double(),
## .. conf_rdt_ov5 = col_double(),
## .. conf_mic_ov5 = col_double()
## .. )
## - attr(*, "problems")=<externalptr>As we did with the vector, we can extract parts of the data frame using indices. For a data frame we can select the nth row, or the nth column using square brackets (note where the comma is paced).
routine_data[,1] # First column## # A tibble: 1,200 × 1
## adm1
## <chr>
## 1 West
## 2 West
## 3 West
## 4 West
## 5 West
## 6 West
## 7 West
## 8 West
## 9 West
## 10 West
## # … with 1,190 more rowsroutine_data[1,] # First row## # A tibble: 1 × 17
## adm1 adm2 hf month year test_u5 test_rdt_u5 test_mic_u5 conf_u5
## <chr> <chr> <dbl> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 West Bamakiary 6 Jan 2018 289 279 127 204
## # … with 8 more variables: conf_rdt_u5 <dbl>, conf_mic_u5 <dbl>,
## # test_ov5 <dbl>, test_rdt_ov5 <dbl>, test_mic_ov5 <dbl>, conf_ov5 <dbl>,
## # conf_rdt_ov5 <dbl>, conf_mic_ov5 <dbl>To access variables of the data frame in base R we can use the
$ operator in the format of
dataframe$variable. We can now look at some of the
variables within our data frame. summary() will provide a
summary of the numerical variable, whilst table() will
tabulate categorical variables.
summary(routine_data$test_u5)## Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
## -9999.00 81.25 136.00 35.13 222.00 702.00 22table(routine_data$adm1)##
## central Central East N. Coast North Coast Plains
## 36 228 276 12 240 144
## West
## 264We can see here that there are -9990 values in the test field. This
is a common way of representing missing data. We can define this as
missing when we import the data in the read_csv
function.
routine_data <- read_csv('data/routine_data.csv', na = c("", "NA", -9999))## Rows: 1200 Columns: 17
## ── Column specification ────────────────────────────────────────────────────────
## Delimiter: ","
## chr (3): adm1, adm2, month
## dbl (14): hf, year, test_u5, test_rdt_u5, test_mic_u5, conf_u5, conf_rdt_u5,...
##
## ℹ Use `spec()` to retrieve the full column specification for this data.
## ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.summary(routine_data$test_u5)## Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
## 0.0 83.0 137.0 164.5 223.0 702.0 37Factors
In our data frame routine_data the variable “month” is a
categorical variable. However, this is currently stored as a
character.
class(routine_data$month)## [1] "character"In R we store and analyse categorical data as factors. It is important that variables are stored correctly so that they are treated as categorical variables in statistical models and data visualisations. We can convert “month” to a factor using this code.
routine_data$month <- factor(routine_data$month)
class(routine_data$month)## [1] "factor"However, month should be an ordered variable and the factor needs to
be ordered accordingly. We can see that R has ordered the factor
alphabetically as standard, we can manually determine the levels of the
factor so that they are ordered correctly using the “levels” option in
the factor() command.
table(routine_data$month)##
## Apr Aug Dec Feb Jan Jul Jun Mar May Nov Oct Sep
## 100 100 100 100 100 100 100 100 100 100 100 100routine_data$month <- factor(routine_data$month,
levels = c("Jan", "Feb", "Mar", "Apr", "May", "Jun",
"Jul", "Aug", "Sep", "Oct", "Nov", "Dec"))
table(routine_data$month)##
## Jan Feb Mar Apr May Jun Jul Aug Sep Oct Nov Dec
## 100 100 100 100 100 100 100 100 100 100 100 100With months we can also achieve this using the helpful specification ‘month.abb’ to tell R we have abbreviated months, rather than listing these out everytime
routine_data$month <- factor(routine_data$month,
levels = month.abb)Task 1
- Open the R project “Data handling and Viz in R”
- Open a new script and save it in the project folder “scripts”
- Install and load the tidyverse packages
- Import the
routine_data.csvfile using theread_csv()function, setting -9999 values to NA, call the object “task_data”- Explore the data using functions such as
str(),head()andsummary()- Convert the variable “month” to an ordered factor
Solution
install.packages("tidyverse") library(tidyverse) routine_data <- read_csv("data/routine_data.csv", na = c('NA', '', -9999)) str(routine_data) head(routine_data) summary(routine_data) routine_data$month <- factor(routine_data$month, levels = month.abb)
Basic data cleaning and manipulation in Tidyverse
In this section we introduce you to the tidyverse
packages and show how these functions can be used to explore, manipulate
and analyse data.
Tidyverse is a collection of R packages for data science, designed to
make cleaning and analyses of data easy and tidy. It uses a range of
packages and functions, along with the pipe operator,
%>%, to produce easily readable and reproducible
code.
We will start by looking at the basic data manipulation functions
from the tidyverse. We have already loaded this package with the command
library(tidyverse).
We will cover the following simple, commonly used functions in tidyverse:
- select
- filter
- rename
- mutate
- group_by and summarise
- drop_na
- arrange and relocate
Additionally, we will cover working with dates in the “lubridate” package and using pipes to combined comands in a clean, stepwise manner
We will continue this investigation using the
routine_data.csv dataset we imported into R in the previous
section.
Select
Firstly, we can use select() to select columns from the
data frame. The first argument is the name of the data frame, followed
by the columns you wish to keep. Here we will just select the tested and
confirmed numbers of malaria cases, along with the location and date
information.
select(routine_data, adm1, adm2, hf, year, month, test_u5, conf_u5)## # A tibble: 1,200 × 7
## adm1 adm2 hf year month test_u5 conf_u5
## <chr> <chr> <dbl> <dbl> <fct> <dbl> <dbl>
## 1 West Bamakiary 6 2018 Jan 289 204
## 2 West Bamakiary 6 2018 Feb 178 92
## 3 West Bamakiary 6 2018 Mar 41 36
## 4 West Bamakiary 6 2018 Apr NA 69
## 5 West Bamakiary 6 2018 May 95 64
....This function can also be used to remove variables from a dataset by using a minus sign before the variables. This means that the same results could be achieved with the following code.
select(routine_data,
-test_rdt_u5, -test_rdt_ov5, -test_mic_u5, -test_mic_ov5,
-conf_rdt_u5, -conf_rdt_ov5, -conf_mic_u5, -conf_mic_ov5)## # A tibble: 1,200 × 9
## adm1 adm2 hf month year test_u5 conf_u5 test_ov5 conf_ov5
## <chr> <chr> <dbl> <fct> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 West Bamakiary 6 Jan 2018 289 204 317 272
## 2 West Bamakiary 6 Feb 2018 178 92 193 104
## 3 West Bamakiary 6 Mar 2018 41 36 45 47
## 4 West Bamakiary 6 Apr 2018 NA 69 137 79
## 5 West Bamakiary 6 May 2018 95 64 101 83
....If we want to assign this to a new object, or replace the existing
object, we can do so using the <- operator.
routine_data <- select(routine_data, adm1, adm2, hf, year, month, test_u5, conf_u5)NOTE: When using tidyverse there are a range of helper functions to
help you concisely refer to multiple variables based on their name. This
makes it easier to select numerous variables and includes helper
functions such as starts_with() and
ends_with(). To explore these further look at
?select
Filter
Next, we can filter the data to only include rows which meet a
certain specification. To do this we use the filter()
function. The first argument is the data frame we wish to filter,
followed by the argument that we want to filter by. In the first command
we select the rows for West region In the second command we select all
rows which tested more than 500 children under 5 for malaria.
We can filter data to be equal to a value using the double equals
sign ==, not equal to using !=, and standard
operators for more than/less than (>, <)
and more/less than or equal to (>=, <=).
Note that when filtering variables that are characters the string must
appear in quotation marks, ““, whilst with numerical values this is not
required.
We can also combine multiple commands into the same filter using the
& (and) or | (or) operators.
filter(routine_data,
adm1 == "West")## # A tibble: 264 × 7
## adm1 adm2 hf year month test_u5 conf_u5
## <chr> <chr> <dbl> <dbl> <fct> <dbl> <dbl>
## 1 West Bamakiary 6 2018 Jan 289 204
## 2 West Bamakiary 6 2018 Feb 178 92
## 3 West Bamakiary 6 2018 Mar 41 36
## 4 West Bamakiary 6 2018 Apr NA 69
## 5 West Bamakiary 6 2018 May 95 64
....filter(routine_data,
test_u5 >500)## # A tibble: 19 × 7
## adm1 adm2 hf year month test_u5 conf_u5
## <chr> <chr> <dbl> <dbl> <fct> <dbl> <dbl>
## 1 West Bamakiary 6 2018 Oct 526 259
## 2 Central Buoadara 65 2018 Sep 527 392
## 3 North Coast Buseli 25 2018 Dec 552 374
## 4 N. Coast Buseli 57 2018 Sep 632 388
## 5 West Cadagudeey 58 2018 Aug 588 339
....filter(routine_data,
(adm1 == "West" | adm1 == "Central") &
test_u5 >=500)## # A tibble: 10 × 7
## adm1 adm2 hf year month test_u5 conf_u5
## <chr> <chr> <dbl> <dbl> <fct> <dbl> <dbl>
## 1 West Bamakiary 6 2018 Oct 526 259
## 2 Central Buoadara 65 2018 Sep 527 392
## 3 West Cadagudeey 58 2018 Aug 588 339
## 4 West Kidobar 95 2018 Nov 502 378
## 5 West Kokam 30 2018 Sep 702 441
....We may want to use the filter function to identify
erroneous data in our dataset, for example, we can identify situations
where the number of people confirmed with malaria is higher than the
number tested.
filter(routine_data,
conf_u5>test_u5)## # A tibble: 19 × 7
## adm1 adm2 hf year month test_u5 conf_u5
## <chr> <chr> <dbl> <dbl> <fct> <dbl> <dbl>
## 1 West Bamakiary 26 2018 Sep 32 68
## 2 N. Coast Buseli 57 2018 Apr 8 9
## 3 North Coast Buseli 98 2018 Jun 24 86
## 4 West Cakure 81 2018 Sep 67 249
## 5 Plains Caya 71 2018 Mar 22 85
....Null values
Null values are treated differently in R. They appear as
NA in the dataset, so we may expect the following code to
work for filtering data to remove all missing values for the number of
people tested for malaria:
filter(routine_data, test_u5 !=NA)However, this does not work as R has a special way of dealing with
missing values. We use the is.na() command, which checks fo
NA values. As with the equals command, if we want the
reverse of this, i.e. “not NA” we can use !is.na(). So the
code to remove missing values would be:
filter(routine_data, !is.na(test_u5))Another method for removing missing data in tidyverse is using the
drop_na() function. As with the filter function this takes
the dataset as the first argument, followed by the variables for which
you are dropping NA values.
routine_data <- drop_na(routine_data, test_u5)Task 2
- Using the “task_data” data frame we imported in the last task:
- Select the variables for location, date and the number of people tested and confirmed cases for children over 5 only (suffix “_ov5”)
- Identify and remove any missing data, or unrealistic values (e.g confirmed cases higher than those tested), overwriting the object “task_data” with this corrected data frame
Solution
task_data <- select(task_data, adm1 , adm2, hf, month, year, test_ov5, conf_u5) task_data <- filter(task_data, !is.na(test_ov5), !is.na(conf_ov5), test_ov5 > conf_ov5)
Rename
There may be situations when we want to rename variables in a data
frame to make it more comprehensive and easier to process. This can
easily be done using the function rename(). We pass to this
function the data frame we are working with, the new name we want the
variable to take and the existing name of the variable. So if we wanted
to change the variable “conf_u5” to “positive_u5”, and to overwrite the
object “routine_data” with this we would simply write:
rename(routine_data,
positive_u5 = conf_u5)## # A tibble: 1,163 × 7
## adm1 adm2 hf year month test_u5 positive_u5
## <chr> <chr> <dbl> <dbl> <fct> <dbl> <dbl>
## 1 West Bamakiary 6 2018 Jan 289 204
## 2 West Bamakiary 6 2018 Feb 178 92
## 3 West Bamakiary 6 2018 Mar 41 36
## 4 West Bamakiary 6 2018 May 95 64
## 5 West Bamakiary 6 2018 Jun 108 42
....Mutate
Next, we can add new variables to the dataset or change existing
variables using mutate(). Mutate allows us to assign new
variables using the = sign. For example, if we wanted to
create a variable of the proportion of tested cases confirmed positive
we could write:
mutate(routine_data,
prop_positive_u5 = conf_u5/test_u5)## # A tibble: 1,163 × 8
## adm1 adm2 hf year month test_u5 conf_u5 prop_positive_u5
## <chr> <chr> <dbl> <dbl> <fct> <dbl> <dbl> <dbl>
## 1 West Bamakiary 6 2018 Jan 289 204 0.706
## 2 West Bamakiary 6 2018 Feb 178 92 0.517
## 3 West Bamakiary 6 2018 Mar 41 36 0.878
## 4 West Bamakiary 6 2018 May 95 64 0.674
## 5 West Bamakiary 6 2018 Jun 108 42 0.389
....We can also use mutate() to alter existing variables
within the data frame. Say we noticed that there was an error in the
admin 1 name where in some instance “North Coast” was entered as “N.
Coast” we can change this by by combining the mutate()
function with an ifelse statement. The first command of the
ifelse() statement is the condition which has to be met,
here adm1 == "N. Coast"; the second command is what you
wish to replace the variable with if this condition is met; the third
part is what the variable should be else this condition is not met.
table(routine_data$adm1)
mutate(routine_data,
country = ifelse(adm1 == "N. Coast", "North Coast", adm1))Similarly, we can use case_when instead of
ifelse when replacing parts of a variable. This is
particularly useful if making multiple changes. As there were other
errors in the adm1 variables we can correct these at the same time using
the following code.
routine_data <-
mutate(routine_data,
adm1 = case_when(adm1 =='N. Coast'~'North Coast',
adm1 == 'central'~'Central',
TRUE~adm1))
table(routine_data$adm1)Dates
Dates in R tidyverse are managed using the lubridate.
This makes working with dates and times in R far easy allowing for
various formats and calculations with dates. In the
lubridate package we can use the following functions to get
the current date and time.
library(lubridate)
today()## [1] "2022-10-23"now()## [1] "2022-10-23 21:07:45 EAT"In our routine_data dataset we have a variable of month and year,
from these we want to create one variable for date. We can do this using
the make_date() function. This function expects inputs for
the day, month and year. If the day or month is missing then this
defaults to 1, and if the year is missing it defaults to 1970.
make_date(year = 2022, month = 6, day = 13)## [1] "2022-06-13"make_date(year = 2022, month = 6)## [1] "2022-06-01"make_date(year = 2022)## [1] "2022-01-01"We can use this function to create a date variable in our routine dataset, combining it with mutate.
routine_data <- mutate(routine_data,
date_tested = make_date(year = year, month = month))Once a date is in the correct date format it is then very easy to
change the format of the date and do calculations on the dates. for more
information look at the lubridate vignette
vignette("lubridate")
Task 3
- Using the “task_data” data frame we imported in the last task:
- Identify any errors in the “year” variable and fix them in the data frame
- Create a new variable containing the date, call this “date_tested”
Solution
table(task_data$year) task_data <- mutate(task_data, year = case_when(year == 18 ~ 2018, year == 3018 ~ 2018), date_tested = make_date(year = year, month = month))
Pipes
We have introduced some of the key functions for data cleaning and
manipulation. However, creating intermediate variables in lots of steps
can be cumbersome. In these cases we can use the pipe operator
%>% to link commands together. This takes the output of
one command and passes it to the next. So if we had the vector called
“tested” we created earlier, and wanted to find the mean of it, rather
than the basic R code we can use the pipe operator to pass the vector
“tested” into the mean function. We can use the shortcut
Ctrl+Shift+M to write the pipe operator.
tested <- c(56, 102, 65, 43, 243)
mean(tested) # Basic command## [1] 101.8tested %>% mean() # Using the pipe## [1] 101.8This means that using our “routine_data” dataset we can link some of the commands we have introduced today together. We can read in our dataset, select the variables we want to keep, filter our dataest, and edit/create variables in one chunk of piped code. Note that as we are using the pipe operator we no longer need to tell each command what data frame is being used.
routine_data <- read.csv("data/routine_data.csv") %>%
select(adm1, adm2, hf, month, year, test_u5, conf_u5) %>%
drop_na(test_u5) %>%
filter(test_u5>conf_u5) %>%
mutate(adm1 = case_when(adm1 =='N. Coast'~'North Coast',
adm1 == 'central'~'Central',
TRUE~adm1),
date_tested = make_date(year = year, month = month))## Warning in make_date(year = year, month = month): NAs introduced by coercionPipes are commonly used when summarising data using the
group_by() and summarise() functions.
Summarising data
There are some useful functions in tidyverse to help you summarise
the data. The first of these is the count() function. This
is a quick function which will allow you to quickly count the
occurrences of an item within a dataset.
count(routine_data, adm1)## adm1 n
## 1 Central 250
## 2 East 262
## 3 North Coast 239
## 4 Plains 138
## 5 West 252By including multiple variables in the command we can count the numbers of times that combination of variables appears.
count(routine_data, adm1, month)## adm1 month n
## 1 Central Apr 22
## 2 Central Aug 20
## 3 Central Dec 21
## 4 Central Feb 22
## 5 Central Jan 22
## 6 Central Jul 22
## 7 Central Jun 20
....If we want to summarise numerical variables we can use the function
summarise(). This is used in conjunction with other
mathematical functions such as sum(), mean(),
median(), max().
summarise(routine_data,
total_tested = sum(test_u5),
mean_tested = mean(test_u5),
median_tested = median(test_u5),
max_tested = max(test_u5))## total_tested mean_tested median_tested max_tested
## 1 190357 166.8335 139 702We can combine the summarise() function with
group_by() to summarise the data by different variables in
the dataset. To calculate the total number of people tested and positive
for malaria in each country in our dataset, we would group by this
variable first and then summarise the data. Grouping is not restricted
to one variable, if you wanted to group the data by location and date
then both variables would be included in the command. Here us an example
of when it is very useful to use the pipe %>%
operator.
clean_ad1_u5 <-
routine_data %>%
group_by(adm1) %>%
summarise(total_tested_u5 = sum(test_u5),
total_positive_u5 = sum(conf_u5))Task 4
- Using the “task_data” data frame from the last task:
- Using the pipe operator group the data by the admin 1 location and the date tested
- Create a new data frame called “summary_data” which contains a summary of the data calculating the minimum, maximum and mean number of confirmed malaria cases in people over 5 by admin 1 location and date tested.
Solution
summary_data <- task_data %>% group_by(adm1, date_tested) %>% summarise(min_cases = min(conf_ov5), mean_cases = mean(conf_ov5), max_cases = max(conf_ov5))
Sorting and reordering data frames
Sorting a data frame by rows and reordering columns is easy in R. To
sort a data frame by a column we use the function
arrange(). We specify the data frame and the column to sort
by, and the default is to sort in ascending order. To sort in a
descending order we can specify this with desc().
Additionally, we can sort by multiple variables, and sorting will be
undertaken in the order they appear in the command.
arrange(routine_data, test_u5)## adm1 adm2 hf month year test_u5 conf_u5 date_tested
## 1 East Galkashiikh 85 Feb 2018 7 5 <NA>
## 2 North Coast Bwiziwo 79 Feb 2018 8 4 <NA>
## 3 East Gakingo 67 Apr 2018 9 4 <NA>
## 4 East Lamanya 7 May 2018 10 8 <NA>
## 5 Central Mbono 41 Jun 2018 10 2 <NA>
## 6 East Yorolesse 54 Feb 2018 10 5 <NA>
## 7 Central Yagoloko 43 Jan 2018 11 7 <NA>
....arrange(routine_data, desc(test_u5))## adm1 adm2 hf month year test_u5 conf_u5 date_tested
## 1 West Kokam 30 Sep 2018 702 441 <NA>
## 2 North Coast Lalaba 47 Nov 2018 661 475 <NA>
## 3 Central Laoye 37 Oct 2018 656 337 <NA>
## 4 North Coast Buseli 57 Sep 2018 632 388 <NA>
## 5 North Coast Lalaba 53 Nov 2018 619 415 <NA>
## 6 East Galkashiikh 5 Sep 2018 608 514 <NA>
## 7 West Cadagudeey 58 Aug 2018 588 339 <NA>
....arrange(routine_data, adm1, date_tested)## adm1 adm2 hf month year test_u5 conf_u5 date_tested
## 1 Central Buoadara 65 Jan 2018 111 106 <NA>
## 2 Central Buoadara 65 Feb 2018 95 50 <NA>
## 3 Central Buoadara 65 Mar 2018 59 32 <NA>
## 4 Central Buoadara 65 Apr 2018 78 43 <NA>
## 5 Central Buoadara 65 May 2018 48 28 <NA>
## 6 Central Buoadara 65 Jun 2018 134 81 <NA>
## 7 Central Buoadara 65 Jul 2018 383 283 <NA>
....We can change the order that columns appear in the dataset using the
relocate() function. This takes the first argument as the
data frame, the second as the column(s) you wish to move, and then where
you want to over them to.
relocate(routine_data, date_tested, .before = adm1)## date_tested adm1 adm2 hf month year test_u5 conf_u5
## 1 <NA> West Bamakiary 6 Jan 2018 289 204
## 2 <NA> West Bamakiary 6 Feb 2018 178 92
## 3 <NA> West Bamakiary 6 Mar 2018 41 36
## 4 <NA> West Bamakiary 6 May 2018 95 64
## 5 <NA> West Bamakiary 6 Jun 2018 108 42
## 6 <NA> West Bamakiary 6 Jul 2018 121 93
## 7 <NA> West Bamakiary 6 Aug 2018 299 175
....Saving data
Once you have finished working on your data there are multiple ways you can save your work in R.
One of the basic ones is to save your dataset as a csv. This is useful as it can easily be opened in other software (such as excel). You may also want to save your data as a Stata (dta) file.
R has some specific formats you can use to store data, these are
.RDS and .RData. RDS files store one R object,
whilst RData can store multiple objects.
Here, we can select which format we want to save the data in and save
the pf_incidence data frame we created in this module.
Similarly to importing data, we can use the base R functions of
write.csv(), or preferably the tidyverse
option if write_csv()
write_csv(clean_ad1_u5, "outputs/clean_admin1_u5.csv")
write_dta(clean_ad1_u5, "outputs/clean_admin1_u5.dta")
saveRDS(clean_ad1_u5, "outputs/clean_admin1_u5.RDS")
save(clean_ad1_u5, population, "outputs/clean_admin1_u5.R")Demo 1
We now want to take everything we have learnt to import and clean the routine dataset. We want our output to contain the total numbers of malaria tests performed and the number of confirmed cases in children under 5, people over 5, and calculate a total for all ages. We want to have the total by admin level 2 locations and for each month in the dataset. This is how we would go about building the code.
routine_data <- read_csv('data/routine_data.csv', na = c("", "NA", -9999))
clean_routine_data <- routine_data %>%
select(adm1, adm2, hf, month, year, test_u5, test_ov5, conf_u5, conf_ov5) %>%
drop_na(test_u5, test_ov5, conf_u5, conf_ov5) %>%
filter(test_u5>conf_u5,
test_ov5>conf_ov5) %>%
mutate(adm1 = case_when(adm1 =='N. Coast'~'North Coast',
adm1 == 'central'~'Central',
TRUE~adm1),
month = factor(month,
levels = month.abb),
year = case_when(year == 3018 ~ 2018,
year == 18 ~ 2018,
TRUE ~ year),
date_tested = make_date(year = year, month = month),
test_total = test_u5+test_ov5,
conf_total = conf_u5+conf_ov5) %>%
group_by(adm1, adm2, date_tested) %>%
summarise(test_total = sum(test_total),
test_u5 = sum(test_u5),
test_ov5 = sum(test_ov5),
conf_total = sum(conf_total),
conf_u5 = sum(conf_u5),
conf_ov5 = sum(conf_ov5))
head(clean_routine_data)## # A tibble: 6 × 9
## # Groups: adm1, adm2 [1]
## adm1 adm2 date_tested test_total test_u5 test_ov5 conf_total conf_u5
## <chr> <chr> <date> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 Central Buoadara 2018-01-01 223 111 112 215 106
## 2 Central Buoadara 2018-02-01 185 95 90 106 50
## 3 Central Buoadara 2018-03-01 120 59 61 75 32
## 4 Central Buoadara 2018-04-01 152 78 74 107 43
## 5 Central Buoadara 2018-05-01 97 48 49 68 28
## 6 Central Buoadara 2018-06-01 272 134 138 188 81
## # … with 1 more variable: conf_ov5 <dbl>write_csv(clean_routine_data, "data/clean_routine_data.csv")Advanced manipulation of data frames
In this section we are building on the what we have introduced in section 2.3 and introducing some more advanced functions for data manipulation. We will be using the “clean_routine_data” dataset we just created.
Reshshaping data
Reshaping or pivoting data is an important part of data cleaning and
manipulation. Tidyverse has introduced the functions
pivot_wider() and pivot_longer() to improve
the ease of reshaping data in R.
pivot_longer() takes a wide dataset and converts it into
a long one, decreasing the number of columns and increasing the number
of rows. Datasets are often created in a wide format, but for analysis a
long format is often preferable, especially for data visualisations.
To reshape the data long we need to pass the argument the columns
which are being pivoted, a name for the new column to identify the
columns being reshaped, and a name for the values of the columns being
reshaped. We can also combine this with helper functions such as
starts_with() to help identify the columns to reshape.
Lets look at an example using the confirmed malaria cases from our
“clean_routine_data” dataset. Here we are taking all of the columns
which start with “conf” and reshaping the data so there is one variable
identifying the age group and one identifying the number of confirmed
cases. We can use the option names_prefix = to identify a
common prefix on the variable names to be removed.
conf_cases <- select(clean_routine_data, adm1, adm2, date_tested, starts_with('conf'))
long_data <- conf_cases %>%
pivot_longer(cols = starts_with("conf"),
names_to = "age_group",
values_to = "confirmed_cases",
names_prefix = "conf_")
head(long_data)## # A tibble: 6 × 5
## # Groups: adm1, adm2 [1]
## adm1 adm2 date_tested age_group confirmed_cases
## <chr> <chr> <date> <chr> <dbl>
## 1 Central Buoadara 2018-01-01 total 215
## 2 Central Buoadara 2018-01-01 u5 106
## 3 Central Buoadara 2018-01-01 ov5 109
## 4 Central Buoadara 2018-02-01 total 106
## 5 Central Buoadara 2018-02-01 u5 50
## 6 Central Buoadara 2018-02-01 ov5 56There are a range of different options in this function to help pivot
the data in the cleanest way possible. To see these you can look at the
vignette by typing the code vignette("pivot").
To pivot the data from long to wide we use the function
pivot_wider(). This is essentially the reverse of the
pivot_longer() function. If we take the object we reshaped
long and want to convert it back to a wide data frame we do this by
specifying the columns which identify the unique observations, and the
columns you wish to reshape. “names_from” is the variable which will
give you the column names, and “values_from” identifies the variable
with the values you want to reshape.
long_data %>% pivot_wider(id_cols = c('adm1', 'adm2', 'date_tested'),
names_from = age_group,
names_prefix = "conf_",
values_from = confirmed_cases) ## # A tibble: 536 × 6
## # Groups: adm1, adm2 [46]
## adm1 adm2 date_tested conf_total conf_u5 conf_ov5
## <chr> <chr> <date> <dbl> <dbl> <dbl>
## 1 Central Buoadara 2018-01-01 215 106 109
## 2 Central Buoadara 2018-02-01 106 50 56
## 3 Central Buoadara 2018-03-01 75 32 43
## 4 Central Buoadara 2018-04-01 107 43 64
....Joining data frames
In R we can easily join two data frames together based on one, or
multiple variables. There are the following options for joining two data
frames, x and y:
inner_join(): includes all rows that appear in both x and yleft_join(): includes all rows in xright_join(): includes all rows in yfull_join(): includes all rows in x or y
To run the command you need to pass it the data frames you wish to join and the variable(s) you wish to join by. If there are matching variables in the data frames these will be detected and you do not need to specify them.
If we have two data frames with varying numbers of rows, we can
investigate the outcomes of using the different join commands. Firstly,
we create two data frames of different lengths, tested, and
confirmed, then look at what the commands and outcomes
would be.
tested <- data.frame(year = c(2015, 2016, 2017, 2018, 2019, 2020),
tested = c(1981, 1992, 2611, 2433, 2291, 2311))
positive <- data.frame(year = c(2013, 2014, 2015, 2016, 2017, 2018),
positive = c(1164, 1391, 981, 871, 1211, 998))
# Command written in full
inner_join(tested, positive, by = "year") ## year tested positive
## 1 2015 1981 981
## 2 2016 1992 871
## 3 2017 2611 1211
## 4 2018 2433 998# Using the pipe operator
tested %>% inner_join(positive) # Keeps only matching records## Joining, by = "year"## year tested positive
## 1 2015 1981 981
## 2 2016 1992 871
## 3 2017 2611 1211
## 4 2018 2433 998tested %>% left_join(positive) # Keeps all records for the first dataset## Joining, by = "year"## year tested positive
## 1 2015 1981 981
## 2 2016 1992 871
## 3 2017 2611 1211
## 4 2018 2433 998
## 5 2019 2291 NA
## 6 2020 2311 NAtested %>% right_join(positive) # Keeps all records for the second dataset ## Joining, by = "year"## year tested positive
## 1 2015 1981 981
## 2 2016 1992 871
## 3 2017 2611 1211
## 4 2018 2433 998
## 5 2013 NA 1164
## 6 2014 NA 1391tested %>% full_join(positive) # Keeps all records from both datasets## Joining, by = "year"## year tested positive
## 1 2015 1981 981
## 2 2016 1992 871
## 3 2017 2611 1211
## 4 2018 2433 998
## 5 2019 2291 NA
## 6 2020 2311 NA
## 7 2013 NA 1164
## 8 2014 NA 1391We can also join datasets which have different names for the variables we wish to join on. Say if the variable for “year” in the positive dataset was “yr”, we could use the command:
positive <- data.frame(yr = c(2013, 2014, 2015, 2016, 2017, 2018),
positive = c(1164, 1391, 981, 871, 1211, 998))
tested %>% inner_join(positive,
by= c("year"="yr"))## year tested positive
## 1 2015 1981 981
## 2 2016 1992 871
## 3 2017 2611 1211
## 4 2018 2433 998Task 5
- Using the “clean_routine_data” data frame from demo 1:
- Import the “clean_routine_data” dataset we created in demo 1 from the “data” folder
- Select the adm1, adm2, date_tested, conf_u5 and conf_ov5 variables
- Pivot this data into the long format so that there is one variable identifying the age group and one identifying the number of confirmed cases.
- Save this data frame as a csv called
confirmed_malariain theoutputsfolder
Solution
clean_data <- read_csv("data/clean_routine_data.csv") clean_data <- select(clean_data, adm1, adm2, date_tested, conf_u5, conf_ov5) clean_data <- > pivot_longer(clean_data, cols = starts_with("conf"), names_to = "age_group", values_to = "confirmed_cases", names_prefix = "conf_"))
Demo 2
In this demo we will link together what we have learnt to create a dataset of the total number of confirmed cases, population and malaria incidence by month, and district. Then another dataset aggergated for the whole country.
# Import the population dataset
pop <- read_csv('data/population.csv')
# Confirmed cases and malaria incidence by age group per month for each admin 2 location
pf_incidence <- inner_join(clean_routine_data, pop) %>%
select(adm1, adm2, date_tested, starts_with('conf'), starts_with('pop') ) %>%
pivot_longer(cols = starts_with("conf")|starts_with("pop"),
names_to = c("metric", "age_group"),
names_sep = "_",
values_to = 'value') %>%
pivot_wider(id_cols = c('adm1', 'adm2', 'date_tested', 'age_group'),
names_from = metric,
values_from = value) %>%
mutate(incidence = conf/pop*100000)
# Confirmed cases and malaria incidence by age group per month for the whole country
pf_incidence_national <-
group_by(pf_incidence,
date_tested, age_group) %>%
summarise(across(c(conf, pop), sum)) %>%
mutate(incidence = conf/pop*100000)
write_csv(pf_incidence, 'outputs/pf_incidence.csv')
write_csv(pf_incidence_national, 'outputs/pf_incidence_national.csv')Introduction to visualization
In this section we will introduce data visualisation using the
popular ggplot package. This package contains functions to
build a range of different plots, from scatter plots and line graphs, to
box plots, bar charts and even maps. It is descriptive and makes it easy
to build on plots and adjust the visualisations to your desired output,
making publication quality plots.
The data we will be using to create our plots is the dataset for the incidence of malaria created from our routine data.
Basic plotting
We will start by showing you how to build a basic plot with the
ggplot() function. The ggplot() function
creates a blank plot to which you add different functions to build your
data visualisation using the + operator.
There are two main arguments to the ggplot() function,
these are:
data: the name of the data frame we are working withmapping: the columns of the data frame we are plotting. This is created using theaes()command.
aes() stands for “aesthetic” and is used to specify what
we want to show in the plot. We will start by looking at plots of simple
time trends using the malaria incidence data for all ages. First, we
filter the pf_incidence_national data frame for
age_group == "total".
total_incidence <-
filter(pf_incidence_national, age_group == 'total')We then start with the ggplot command. We want to use the total incidence dataset and wish to plot the year on the x-axis and the incidence on the y-axis so the command is as follows:
ggplot(data = total_incidence, mapping = aes(x = date_tested, y = incidence))Scatter plots & line graphs
This wont yet produce a figure as we need to tell ggplot what sort of
figure we would like this to be. We will start by making a scatter plot
using geom_point(). We add layers to the ggplot object
using the + operator.
ggplot(data = total_incidence, mapping = aes(x = date_tested, y = incidence))+
geom_point()
We can then control how the points look by adding commands to the
geom_point() function, such as the size, colour and
transparency (alpha) of the points.
ggplot(data = total_incidence, mapping = aes(x = date_tested, y = incidence))+
geom_point(size=5, colour = "blue", alpha = 0.5)
We can also change the type of the point by using the
shape = command.
ggplot(data = total_incidence, mapping = aes(x = date_tested, y = incidence))+
geom_point(size=4, colour = 'blue', alpha = 0.5, shape = 18)
# Available shapes (don't run)
shapes
The general structure of the ggplot() command remains
similar for different plots.This means that it is easy to change the
type of an established plot, or create a different plot using a similar
code structure.
We can change the type of plot by replacing geom_point()
with other options such as a line graph: geom_line(), or
smoothed conditional means: geom_smooth().
# Line plot
ggplot(data = total_incidence, mapping = aes(x = date_tested, y = incidence))+
geom_line(size=2, colour = "red", alpha = 0.5)
# Smoothed line of fit
ggplot(data = total_incidence, mapping = aes(x = date_tested, y = incidence))+
geom_smooth(colour = "yellow")

The size, colour and alpha commands are similarly used to control the
appearance of the line but instead of using shape = to
control the appearance of the points we can change the style of the line
in a line plot by specifying linetype =
ggplot(data = total_incidence, mapping = aes(x = date_tested, y = incidence))+
geom_line(size=2, colour = "red", alpha = 0.5, linetype = 'dashed')
# Available line types (not run)
lty

Additionally, you can layer the types of plots together, e.g. plotting the points and lines on the same graph.
ggplot(data = total_incidence, mapping = aes(x = date_tested, y = incidence))+
geom_line(size=1, colour = "red", alpha = 0.5)+
geom_point(size=5, colour = 'blue', shape = 18)
Task 6
- Import the dataset
pf_incidence_nationalwe created in the demo from the data folder and filter to just contain data for under 5’s- Create a line graph plotting the incidence of malaria (y-axis) by date (x-axis)
- Change this to be a green dotted line and increase the line thickness
Solution
pf_incidence_national <- read_csv("outputs/pf_incidence_national.csv") %>% filter(age_group=="u5") ggplot(pf_incidence_national)+ geom_line(mapping = aes(x = date_tested, y = incidence), color = 'green', size = 2)
Themes
Further commands can then be added to the ggplot() object to control
the appearance of the plot. We can label the axis and add a title using
labs(), and can select different themes for the background
appearance, such as the black and white theme: theme_bw(),
or the dark theme: theme_dark().
ggplot(data = total_incidence, mapping = aes(x = date_tested, y = incidence))+
geom_point(size=2, colour = "blue")+
labs(x = "Date",
y = "Incidence/100,000/month",
title = "Malaria incidence in 2018")+
theme_bw()
We can control the x and y axis limits by using xlim()
or ylim(). As our x-axis is a date we can control this
using the command scale_x_date(). Otherwise if this was a
continuous variable we could use scale_x_continous() to
control the scale. This command has additional options to control where
you want the breaks in the axis to be and to alter the data labels
# Option 1 using xlim() and ylim()
ggplot(data = total_incidence, mapping = aes(x = date_tested, y = incidence))+
geom_point(size=5, colour = 'blue')+
labs(x = "Date",
y = "Incidence/100,000/month",
title = "Malaria incidence in 2018")+
theme_bw()+
ylim(0,1000)
ggplot(data = total_incidence, mapping = aes(x = date_tested, y = incidence))+
geom_point(size=5, colour = 'blue')+
labs(x = "Date",
y = "Incidence/100,000/month",
title = "Malaria incidence in 2018")+
theme_bw()+
scale_x_date(limit=c(as.Date("2018-01-01"),as.Date("2018-06-01")),
date_breaks = "1 month", date_labels = "%b")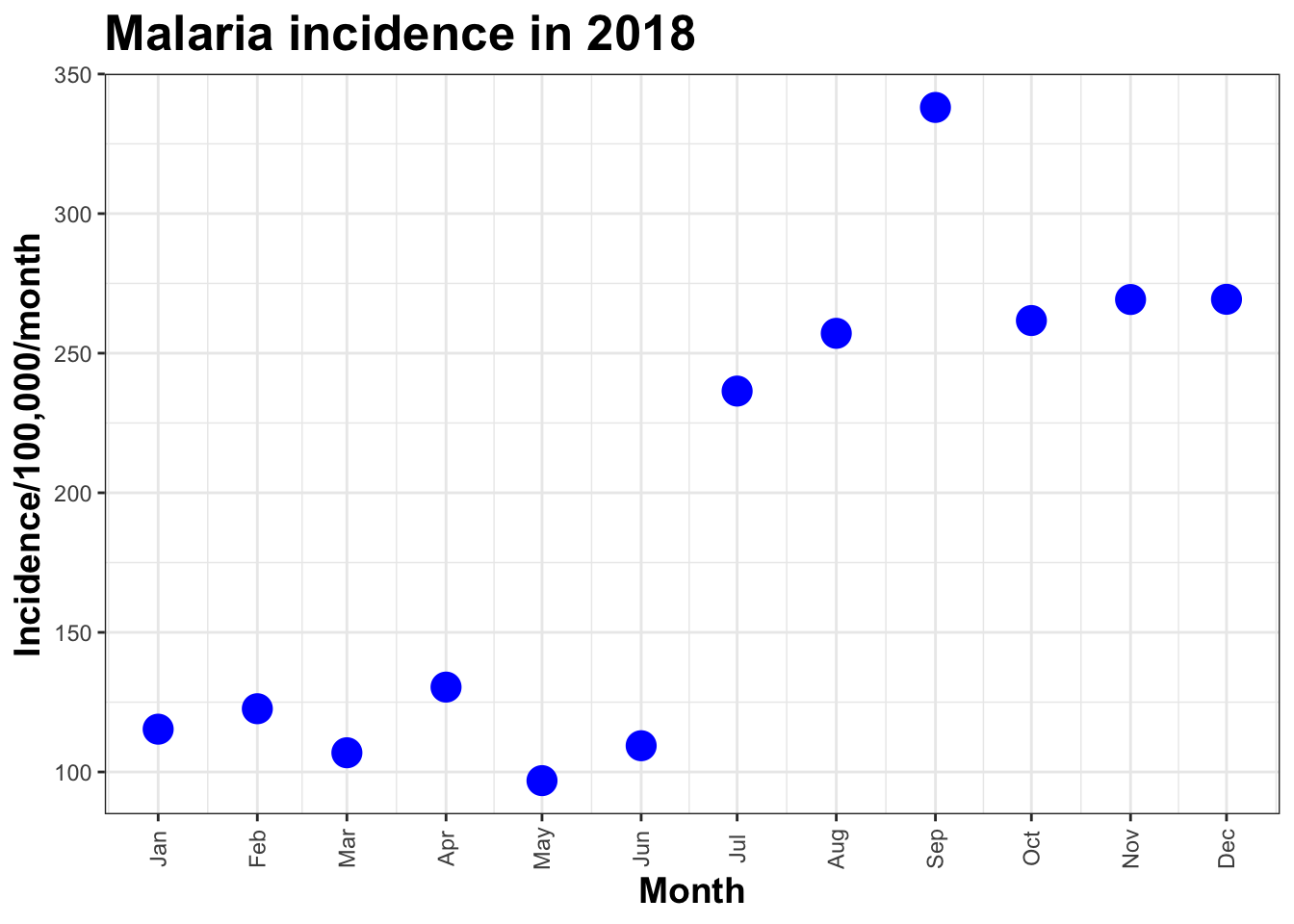

Furthermore, we can control the text of the axis and title within the
theme() command. theme() provides a large
amount of control over the appearance of the plot. Common examples of
what we can do within this command is rotating the x-axis text using the
option angle =, changing the font size using
size = and the font type using face =.
ggplot(data = total_incidence, mapping = aes(x = date_tested, y = incidence))+
geom_point(size=5, colour = 'blue')+
labs(x = "Month",
y = "Incidence/100,000/month",
title = "Malaria incidence in 2018")+
theme_bw()+
scale_x_date(date_breaks = "1 month", date_labels = "%b")+
theme(axis.text.x = element_text(angle = 90, hjust = 0.5, vjust = 0.5),
axis.title.x = element_text(size = 14,face = 'bold'),
axis.title.y = element_text(size = 14,face = 'bold'),
title = element_text(size = 16,face = 'bold'))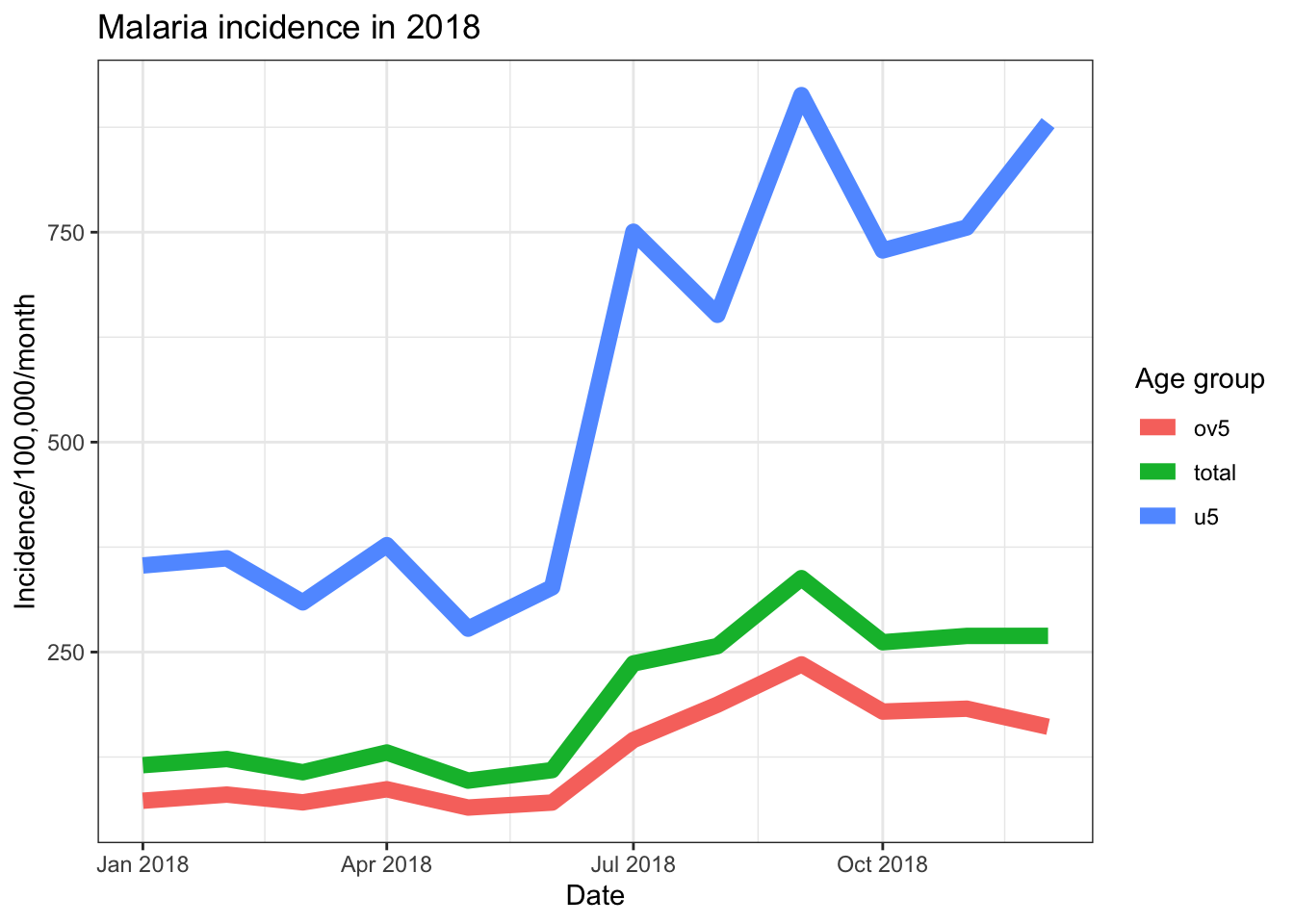
Subgroups and facets
We will now look visualising multiple data trends, for example if you
had incidence data for multiple countries there are various ways you
could show this data. Here we will use the dataset
pf_incidence_national.csv we imported earlier, with
incidence for different age groups.
Firstly, we can incorporate the trends for different age_groups on
the same plot by using different colours. We use the colour
argument inside the aes functions and assign it to the
country variable. This results in ggplot plotting a line for each
country in the dataset in a different colour, with a legend added to the
plot.
ggplot(data = pf_incidence_national, mapping = aes(x = date_tested, y = incidence, colour = age_group))+
geom_line(size = 3)+
labs(x = "Date",
y = "Incidence/100,000/month",
title = "Malaria incidence in 2018",
colour = 'Age group')+
theme_bw()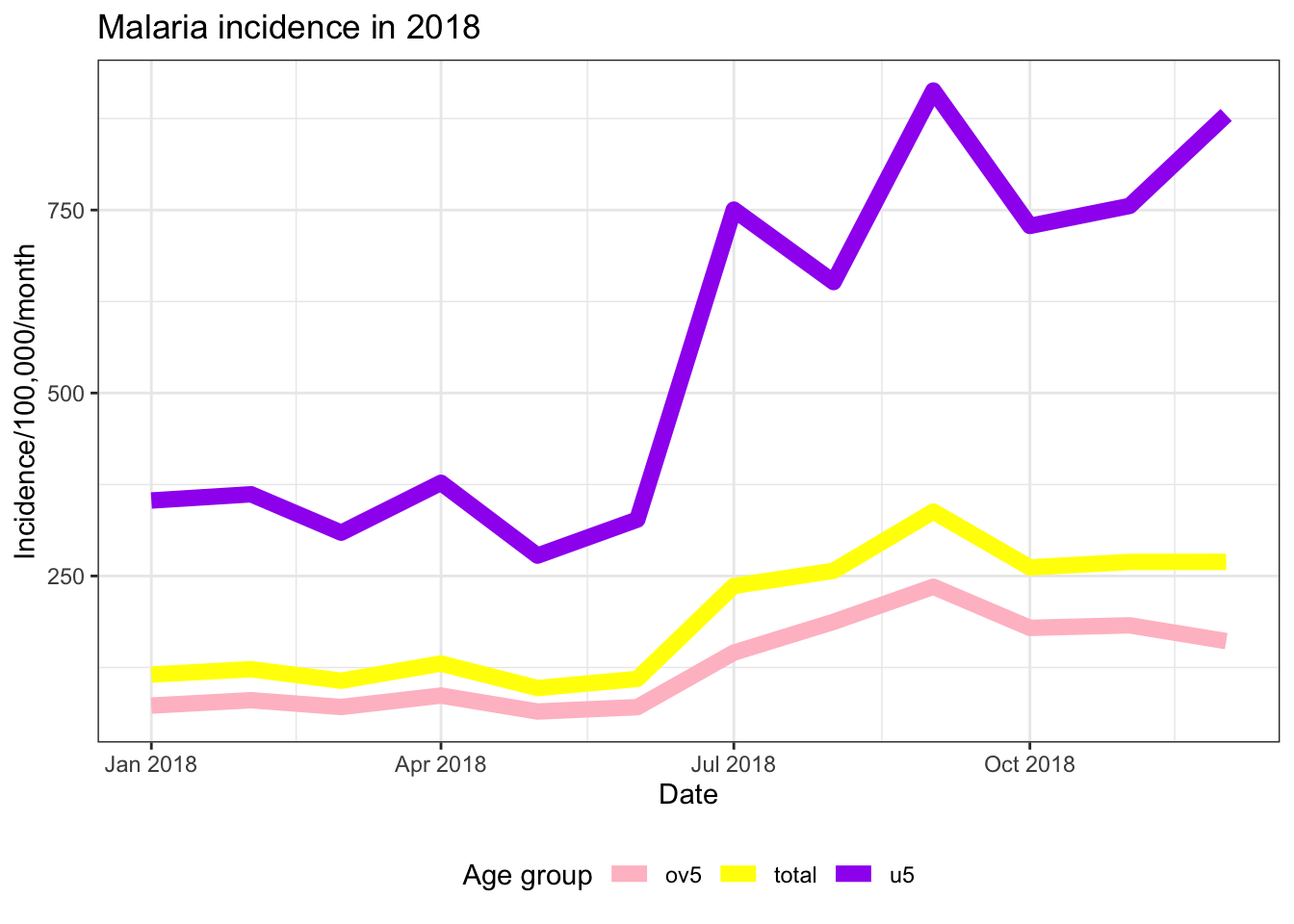
We can control the colours in the plots by using the
scale_color_manual() function, and passing it the colours
you wish to use. You can use either the names of colours or the HEX
codes. We can also move the position of the legend to the bottom using
the legend.position = "bottom" command within
theme() and change the title of the legend within
labs().
ggplot(data = pf_incidence_national, mapping = aes(x = date_tested, y = incidence, colour = age_group))+
geom_line(size = 3)+
labs(x = "Date",
y = "Incidence/100,000/month",
title = "Malaria incidence in 2018",
colour = 'Age group')+
theme_bw()+
theme(legend.position = 'bottom')+
scale_colour_manual(values = c('pink', 'yellow', 'purple'))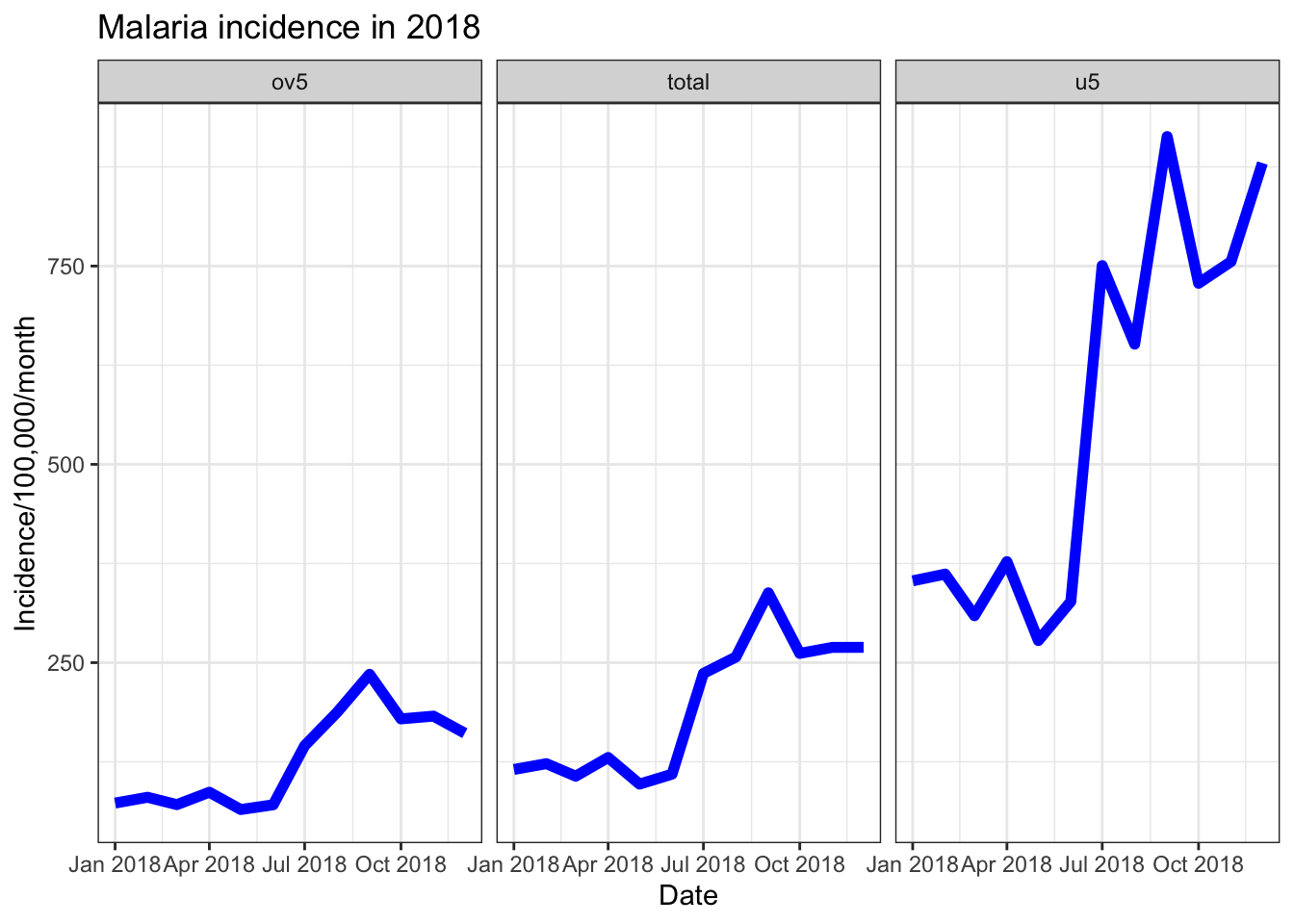
Another way to look at multiple data trends in one plot is to uses facets. This creates separate plots for each “facet”, as part of the overall plot.
ggplot(data = pf_incidence_national, mapping = aes(x = date_tested, y = incidence))+
geom_line(size=2, colour = "blue")+
labs(x = "Date",
y = "Incidence/100,000/month",
title = "Malaria incidence in 2018")+
theme_bw()+
facet_wrap(~age_group)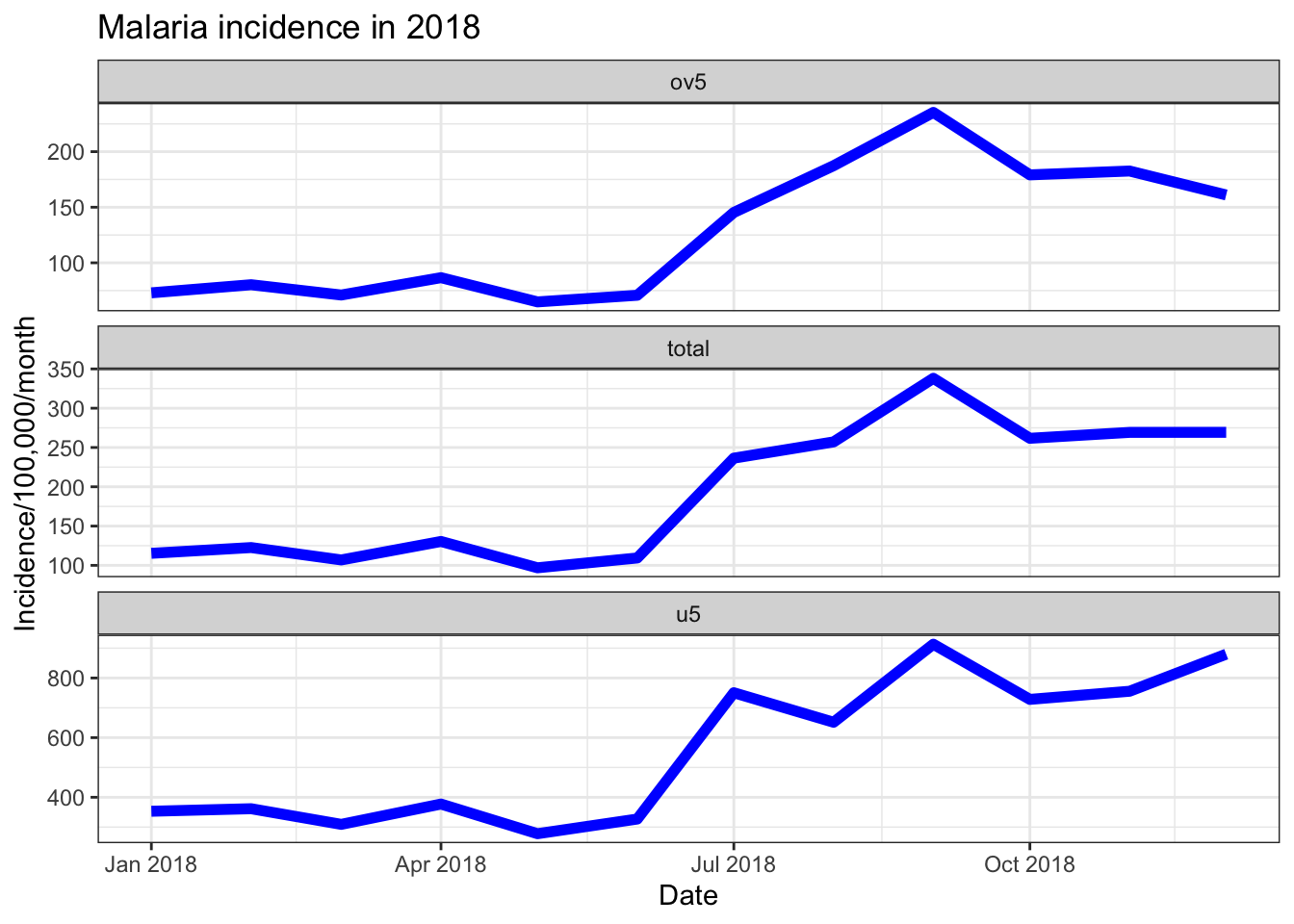
There are various options in facet_wrap(). If you have
largely varying scales between the facets you are plotting, you can plot
these on different scales using either free_y = TRUE, or
free_x = TRUE as options inside of
facet_wrap(). You can also specify the number of rows or
columns you want in the plot, e.g. if you had 6 facets you could plot it
as nrow = 2, ncol = 3 or
nrow = 3, ncol = 2.
ggplot(data = pf_incidence_national, mapping = aes(x = date_tested, y = incidence))+
geom_line(size=2, colour = "blue")+
labs(x = "Date",
y = "Incidence/100,000/month",
title = "Malaria incidence in 2018")+
theme_bw()+
facet_wrap(~age_group, scales = 'free_y', nrow = 3)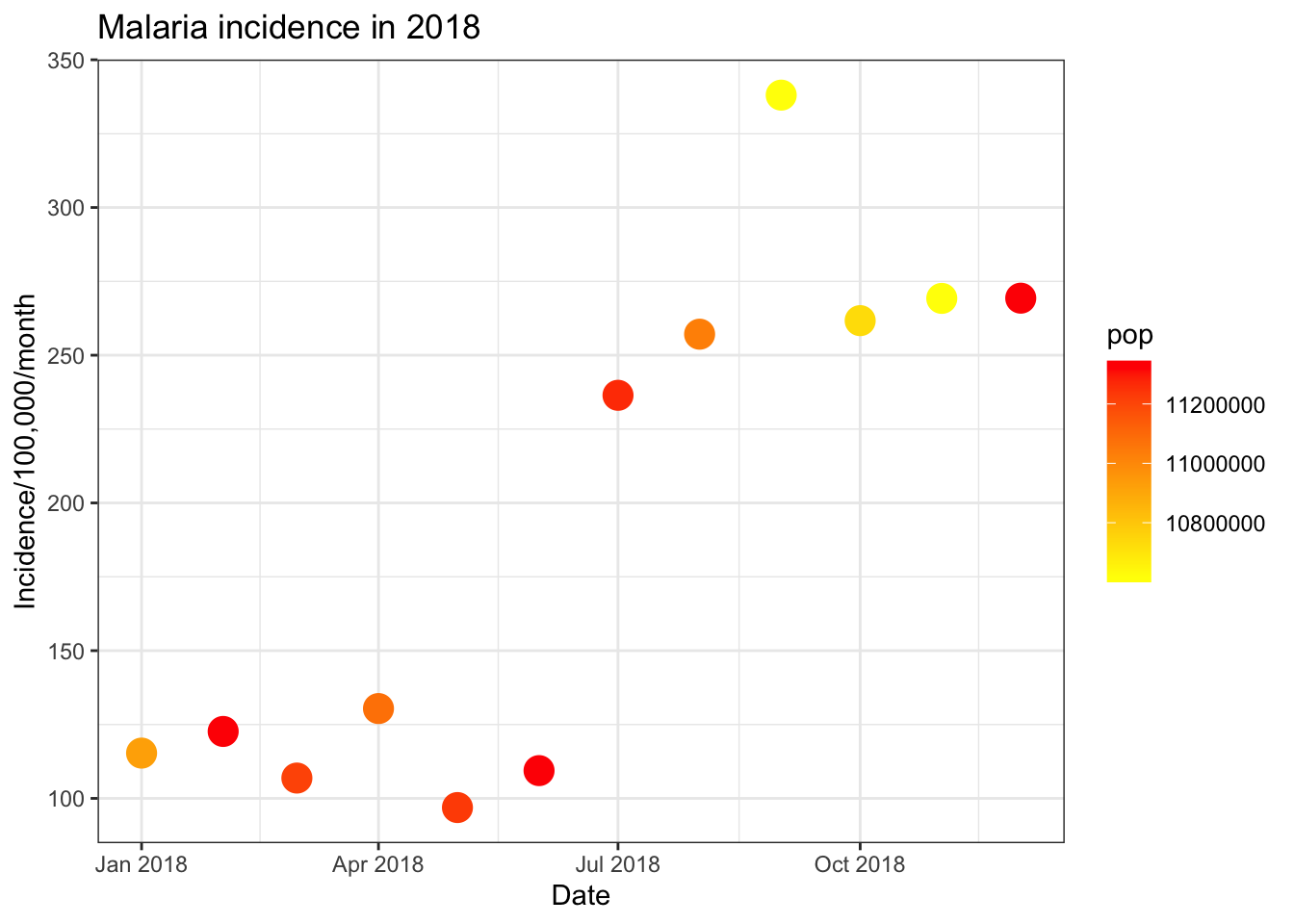
Task 7
- Using the
pf_incidence_nationaldataset create scatter plots the number of confirmed malaria cases(y-axis) by date tested (x-axis), with facets for age group.- Change the colour of the points based on the age group
- Change the axis labels and add a title to the plot
- Change the plot theme to black and white
Solution
pf_incidence_national <- read_csv("outputs/pf_incidence_national.csv") ggplot(pf_incidence_national)+ geom_line(mapping = aes(x = date_tested, y = incidence, color = >age_group))+ facet_wrap(~age_group)+ labs(x = "Date", y = "Incidence per 1000 PYO", title = "Incidence >Rate")+ theme_bw()
Continuous colour scales
Similarly to using the colour option to plot your data
by categorical variables, we can use this options to plot continuous
data on a colour scale. For example, if we wanted colour the points on
our plot of incidence by year based on the total population we can add
colour = population to the aesthetic command.
ggplot(total_incidence, aes(x = date_tested, y = incidence, colour = pop))+
geom_point(size=5)+
labs(x = "Date",
y = "Incidence/100,000/month",
title = "Malaria incidence in 2018")+
theme_bw()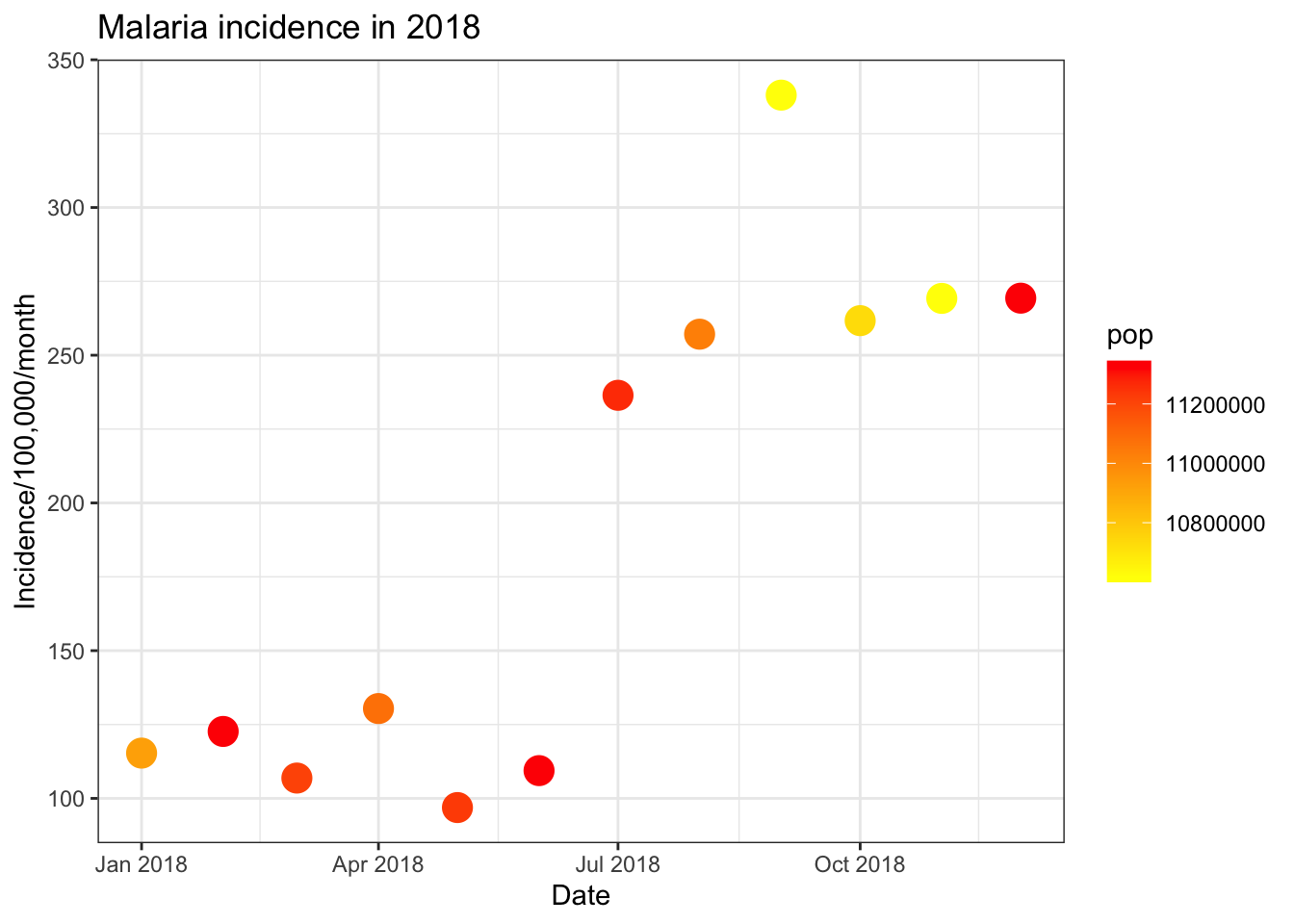
There are then a few options for how we can control the colour scale
of a continuous variable. Firstly we can use the
scale_colour_gradient() command to specify the low and high
colours of a gradient from which to build the colour ramp. Secondly
ggpplot allows us to use colour scales from ColorBrewer (https://colorbrewer2.org). This has a wide range of
sequential, diverging and qualitative colour schemes you can use for a
rnage of data and projects. For continuous data this is specified using
the command scale_colour_distiller() and using the option
palette = to specify the colour palette to use.
ggplot(total_incidence, aes(x = date_tested, y = incidence, colour = pop))+
geom_point(size=5)+
labs(x = "Date",
y = "Incidence/100,000/month",
title = "Malaria incidence in 2018")+
theme_bw()+
scale_colour_gradient(low = "yellow", high = "red")
ggplot(total_incidence, aes(x = date_tested, y = incidence, colour = pop))+
geom_point(size=5)+
labs(x = "Date",
y = "Incidence/100,000/month",
title = "Malaria incidence in 2018")+
theme_bw()+
scale_colour_distiller(palette = 'Greens')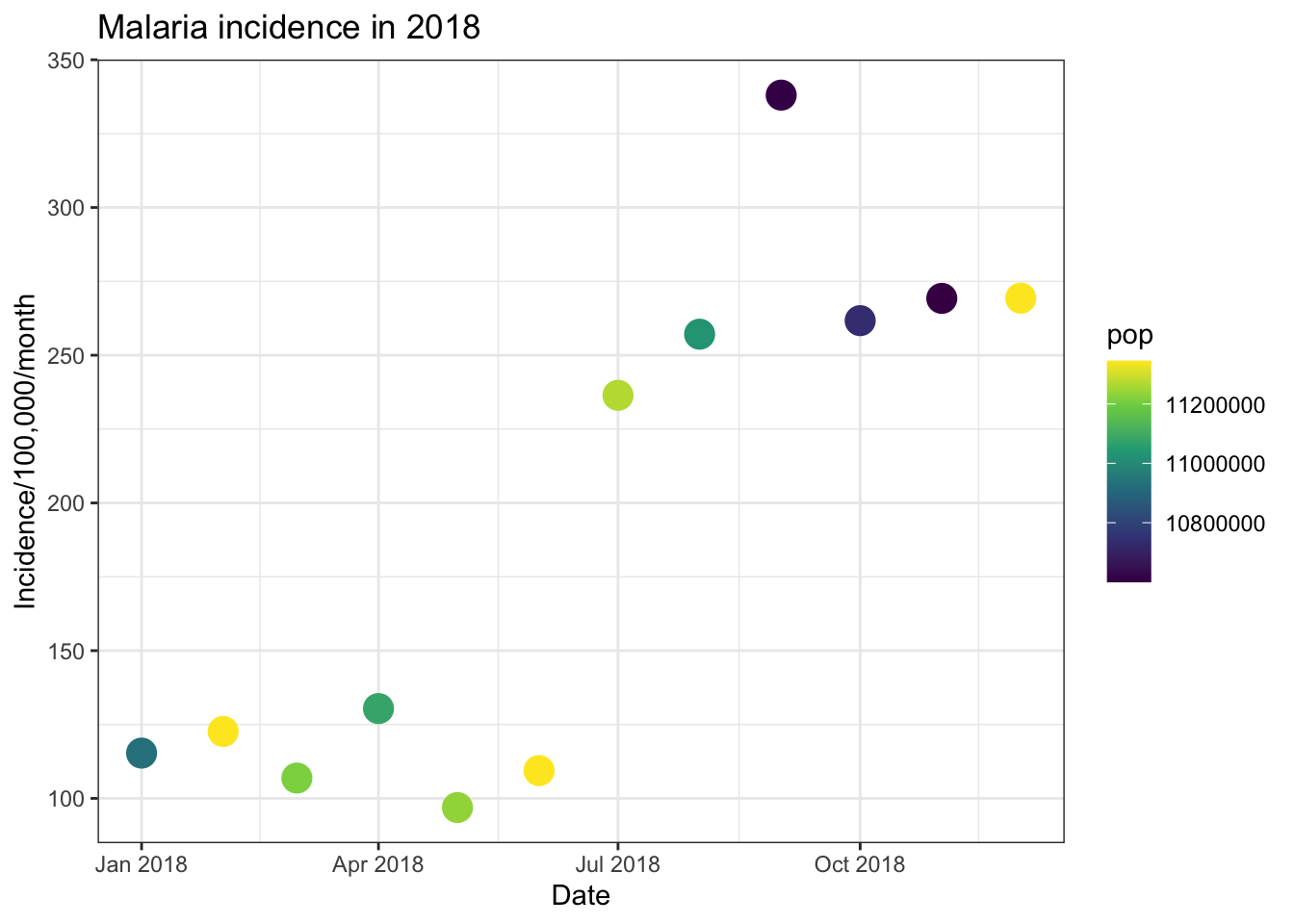
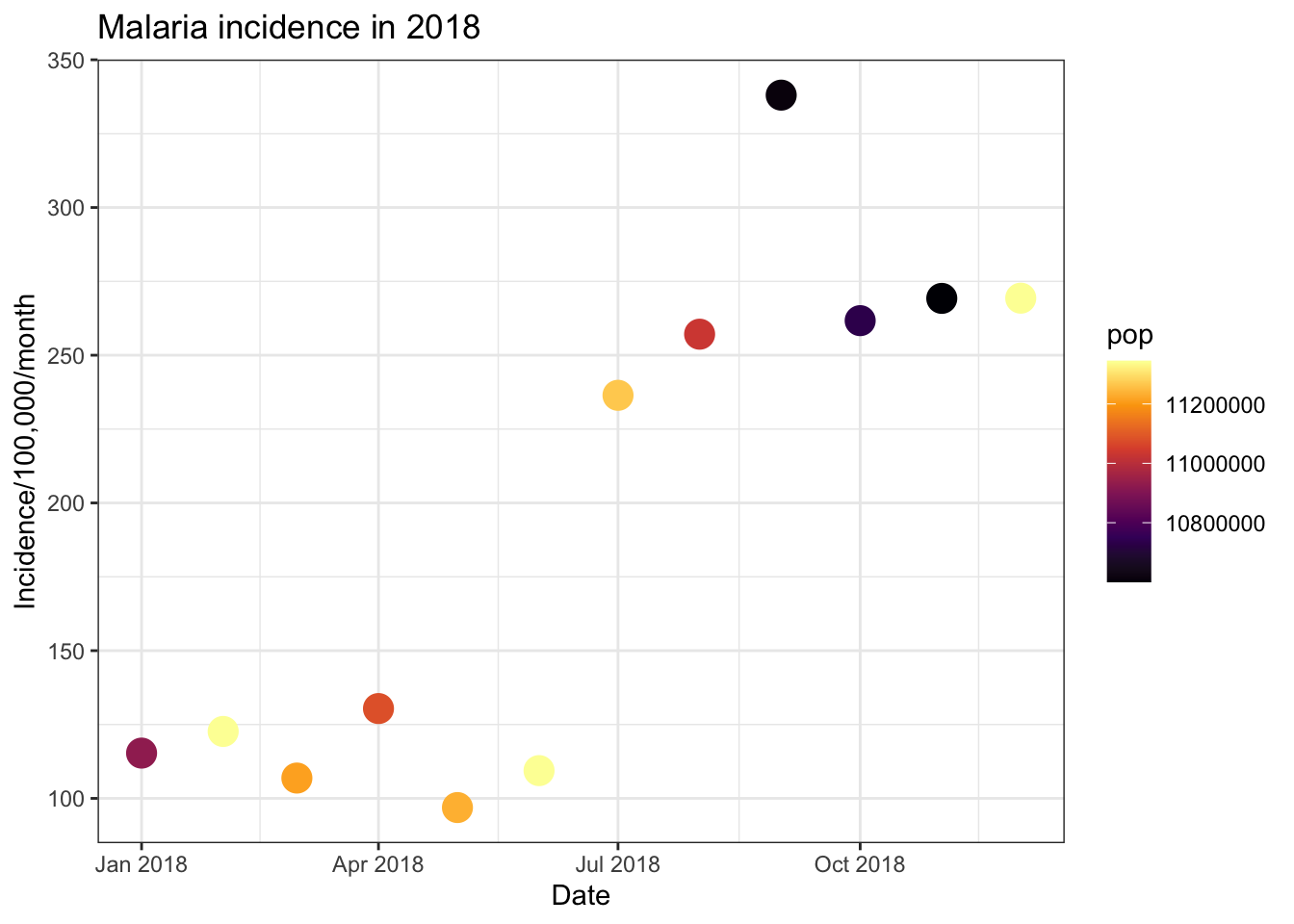
An additional option for- continuous colour scales are the viridis
scales. These are well designed colour scales which are excellent for
graphs and maps. They are colourful, appear uniform in both colour and
black-and-white and are easy to view for persons with common forms of
colour blindness. There are various options withing this package and
they can be used within ggplot through the command
scale_colour_viridis_c().
# Default viridis colour scale
ggplot(total_incidence, aes(x = date_tested, y = incidence, colour = pop))+
geom_point(size=5)+
labs(x = "Date",
y = "Incidence/100,000/month",
title = "Malaria incidence in 2018")+
theme_bw()+
scale_colour_viridis_c()
# Inferno option
ggplot(total_incidence, aes(x = date_tested, y = incidence, colour = pop))+
geom_point(size=5)+
labs(x = "Date",
y = "Incidence/100,000/month",
title = "Malaria incidence in 2018")+
theme_bw()+
scale_colour_viridis_c(option = 'inferno')
# Reversing the direction
ggplot(total_incidence, aes(x = date_tested, y = incidence, colour = pop))+
geom_point(size=5)+
labs(x = "Date",
y = "Incidence/100,000/month",
title = "Malaria incidence in 2018")+
theme_bw()+
scale_colour_viridis_c(option = 'inferno', direction = -1)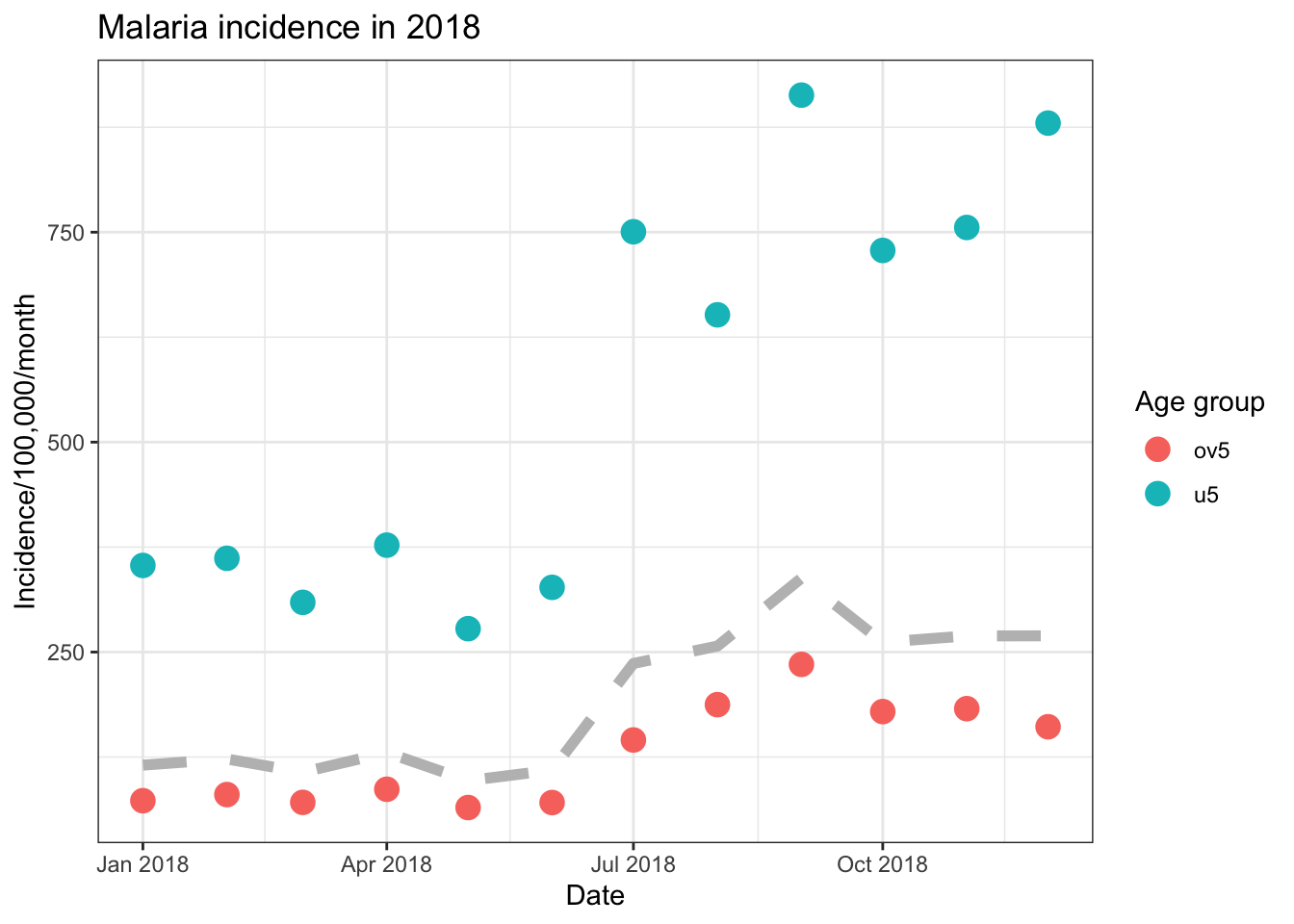


Finally, we can use ggplot to have plot different data on different
layers of the plot. We do this by moving the data = and
mapping = arguments from the ggpplot() to the
specific layers such as geom_point(). Here we plot a
scatter plot with different colours for each country, but we add a
smoothed mean line for all of the data.
ggplot()+
geom_point(data = filter(pf_incidence_national, age_group != 'total'),
aes(x = date_tested, y = incidence, colour = age_group), size=4)+
geom_line(data = filter(pf_incidence_national, age_group == 'total'),
aes(x = date_tested, y = incidence), colour = 'grey', size = 2, linetype = 'dashed')+
labs(x = "Date",
y = "Incidence/100,000/month",
title = "Malaria incidence in 2018",
colour = 'Age group')+
theme_bw()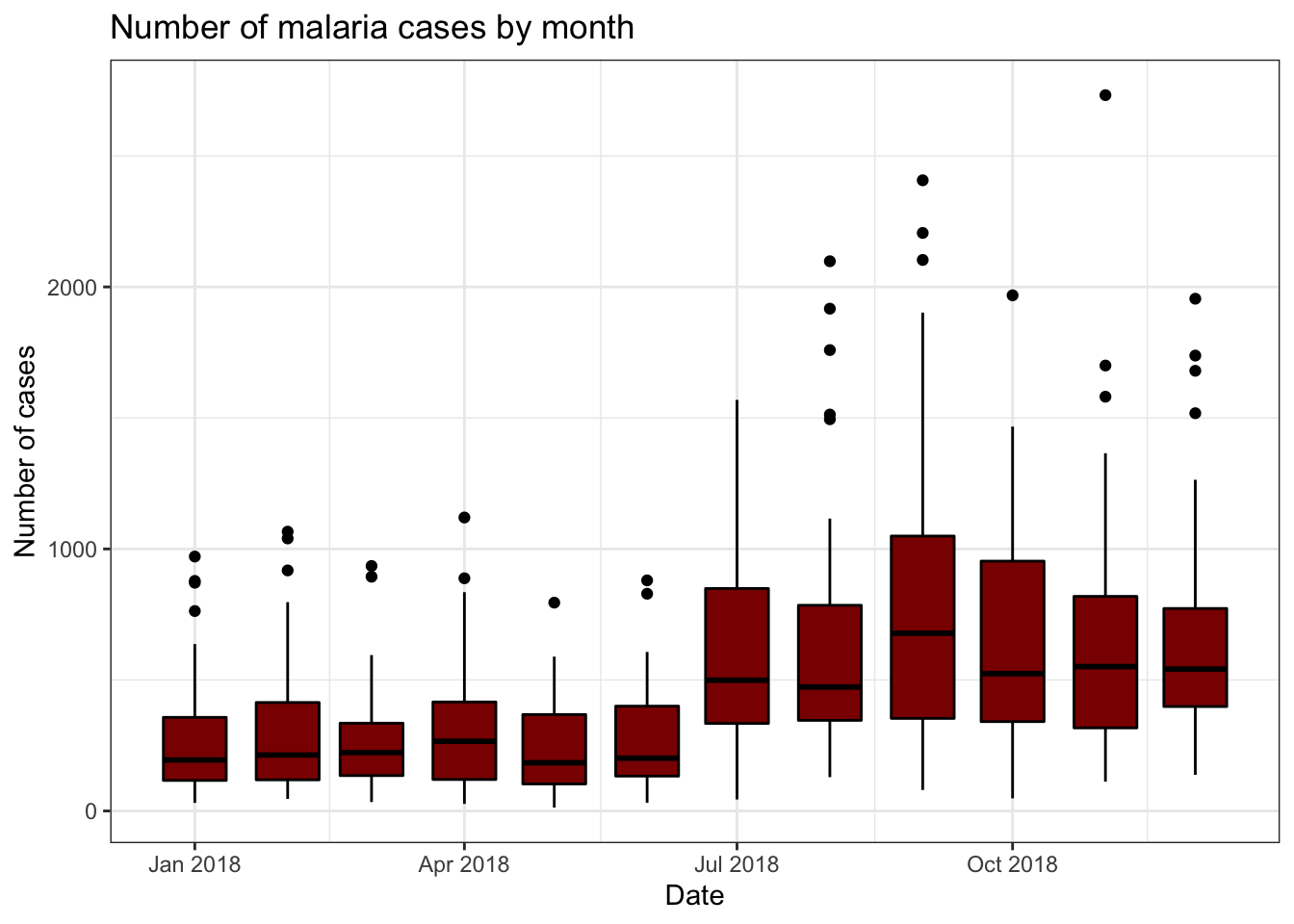
Box plots, bar charts and histograms
ggplot can also be used to create plots summarising the
data and incorporating statistical transformations.
Box plots are a good way of summarising continuous data by discrete
variables. For example, in this dataset we have the number of confirmed
malaria cases in different districts for each month. In
ggplot we can use box plots to easiy summarise and
visualise these data.
As in the previous section, we provide ggplot() with the x and y
variables in the aes() argument and R will calculate the
size of the boxes and whiskers. Similarly, you can control the colour of
the lines and the fill colour of the plot by using the
colour and fill arguments.
To look at these plots we will use the “pf_incidence” dataset we
created earlier, with the confirmed cases, incidence, and population by
date and district. We will subset this data to look at all ages
initially. We introduced pipes earlier, these can also be used with
ggplot. Dates in ggplot are treated as continuous variables to here we
must specify group = date_tested to group the data by
date.
filter(pf_incidence, age_group == 'total') %>%
ggplot(aes(x = date_tested, y = conf, group=date_tested))+
geom_boxplot(colour = 'black', fill = 'dark red')+
labs(x = "Date",
y = "Number of cases",
title = "Number of malaria cases by month")+
theme_bw()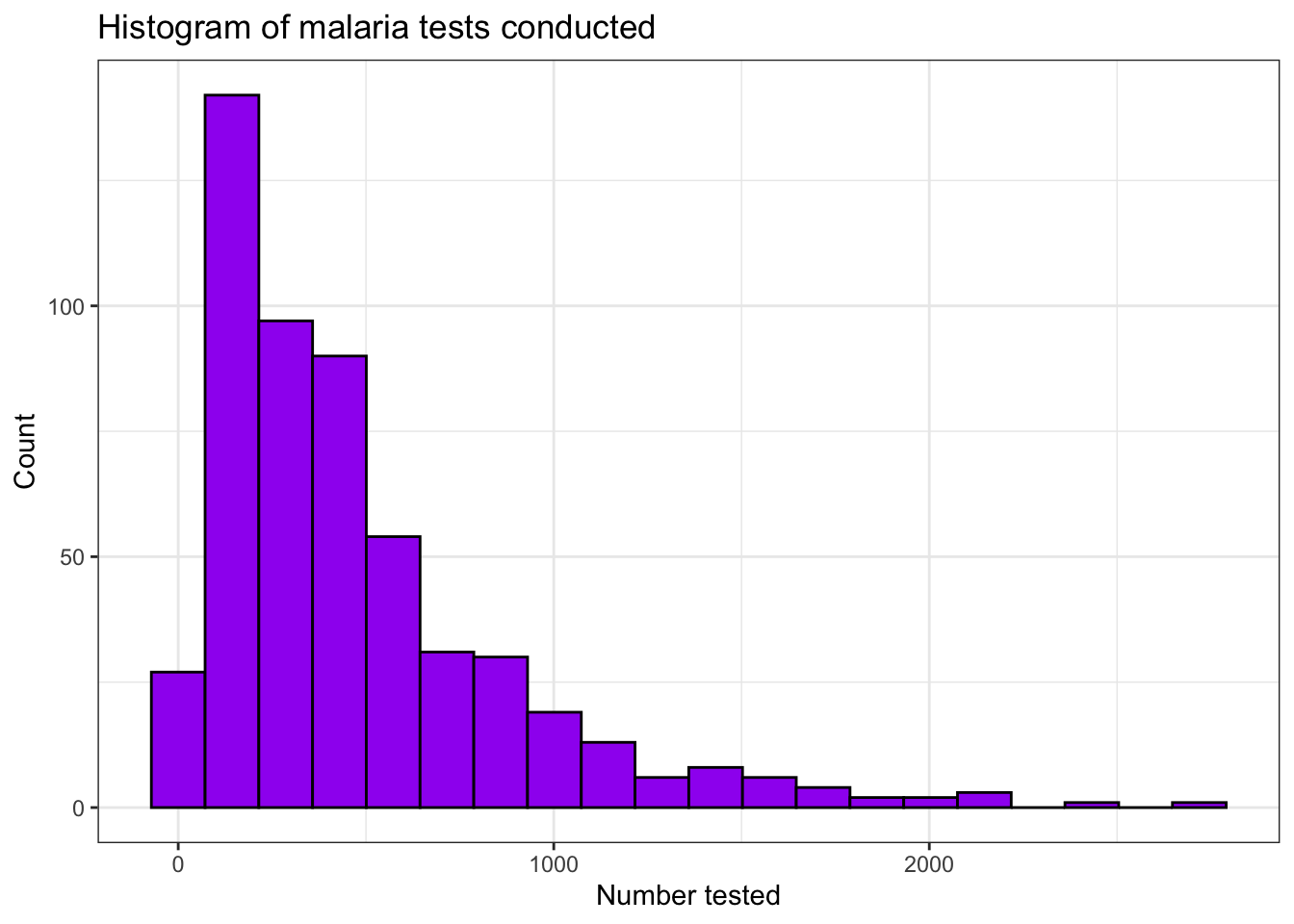
Histograms are created using the geom_histogram()
function. This splits the data into bins using stat_bins()
and counts the number of occurrences in each bin. You can control the
number of bins or the width of the bins using bins and
binwidth respectively. Options such as fill
can also be used to control the fill colour of the bars, along with the
additional options already discussed.
filter(pf_incidence, age_group == 'total') %>%
ggplot(aes(x = conf))+
geom_histogram(bins = 20, fill = 'purple', colour = 'black')+
theme_bw()+
labs(x = "Number tested",
y = "Count",
title = "Histogram of malaria tests conducted")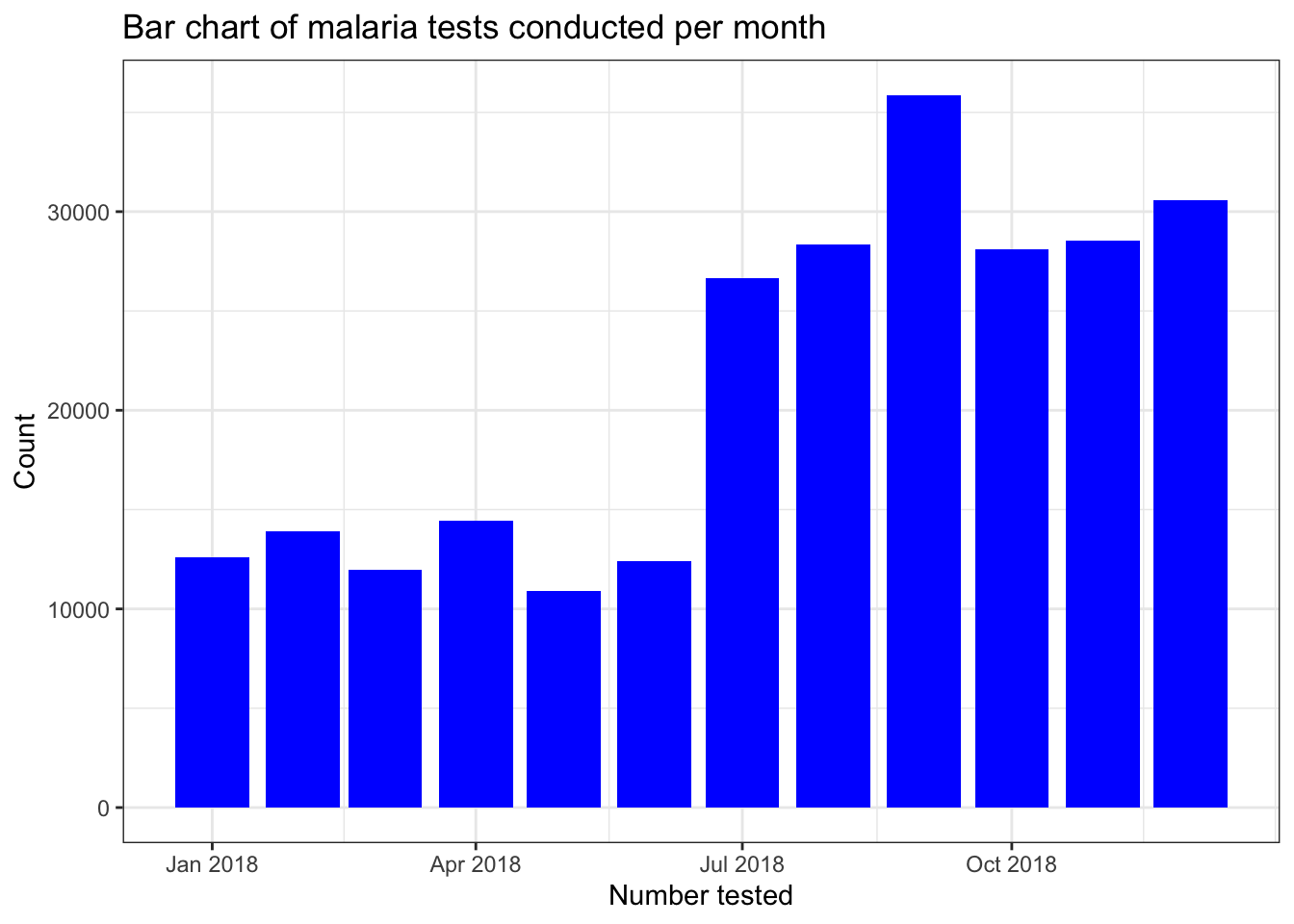
For bar plots the geom_bar()function is used. The
default transformation is to count the number of rows for each category,
stat = "bin". There are other transformations we can use,
so if we want to make the heights of the bars to represent values in the
data (provided by the assigning the y aesthetic), we use
stat = identity. Here we plot a bar chart of the total
number of malaria tests conducted each month in our routine data.
filter(pf_incidence, age_group == 'total') %>%
ggplot(aes(x = date_tested, y = conf, group=date_tested))+
geom_bar(stat = "identity", fill = "blue")+
labs(x = "Number tested",
y = "Count",
title = "Bar chart of malaria tests conducted per month") +
theme_bw()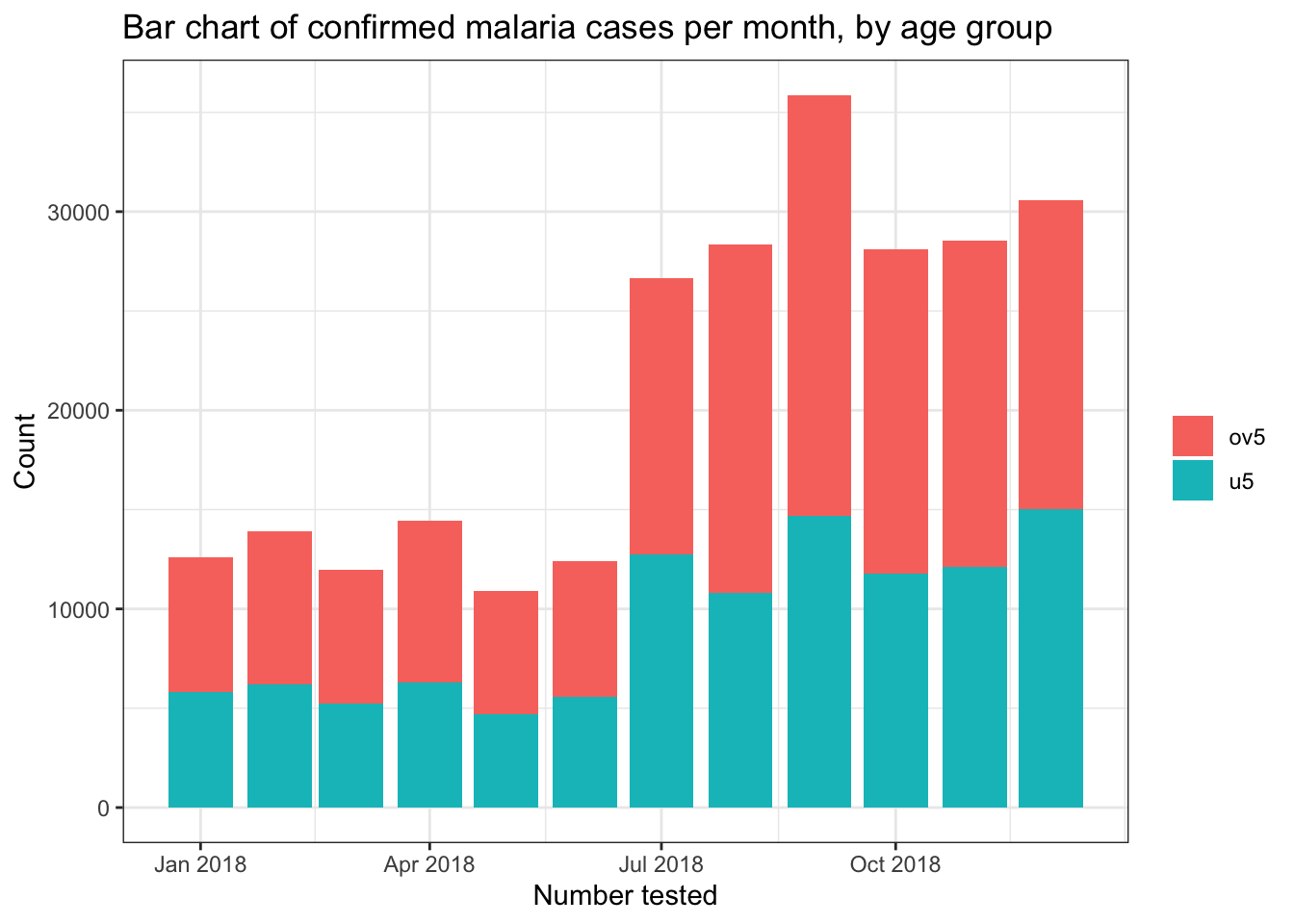
If we want to display different subgroups of data, such as number of
tests by age group, on a bar plot we can either create a stacked bar
plot with different colours representing the subgroups by using the
fill = age_group command, or by creating a side-by-side bar
plot by combining fill = age_group with the command
position = "dodge".
# Stacked bar chart
filter(pf_incidence, age_group != 'total') %>%
ggplot(mapping = aes(x = date_tested, y = conf, fill = age_group))+
geom_bar(stat = "identity")+
theme_bw()+
labs(x = "Number tested",
y = "Count",
title = "Bar chart of confirmed malaria cases per month, by age group",
fill = NULL)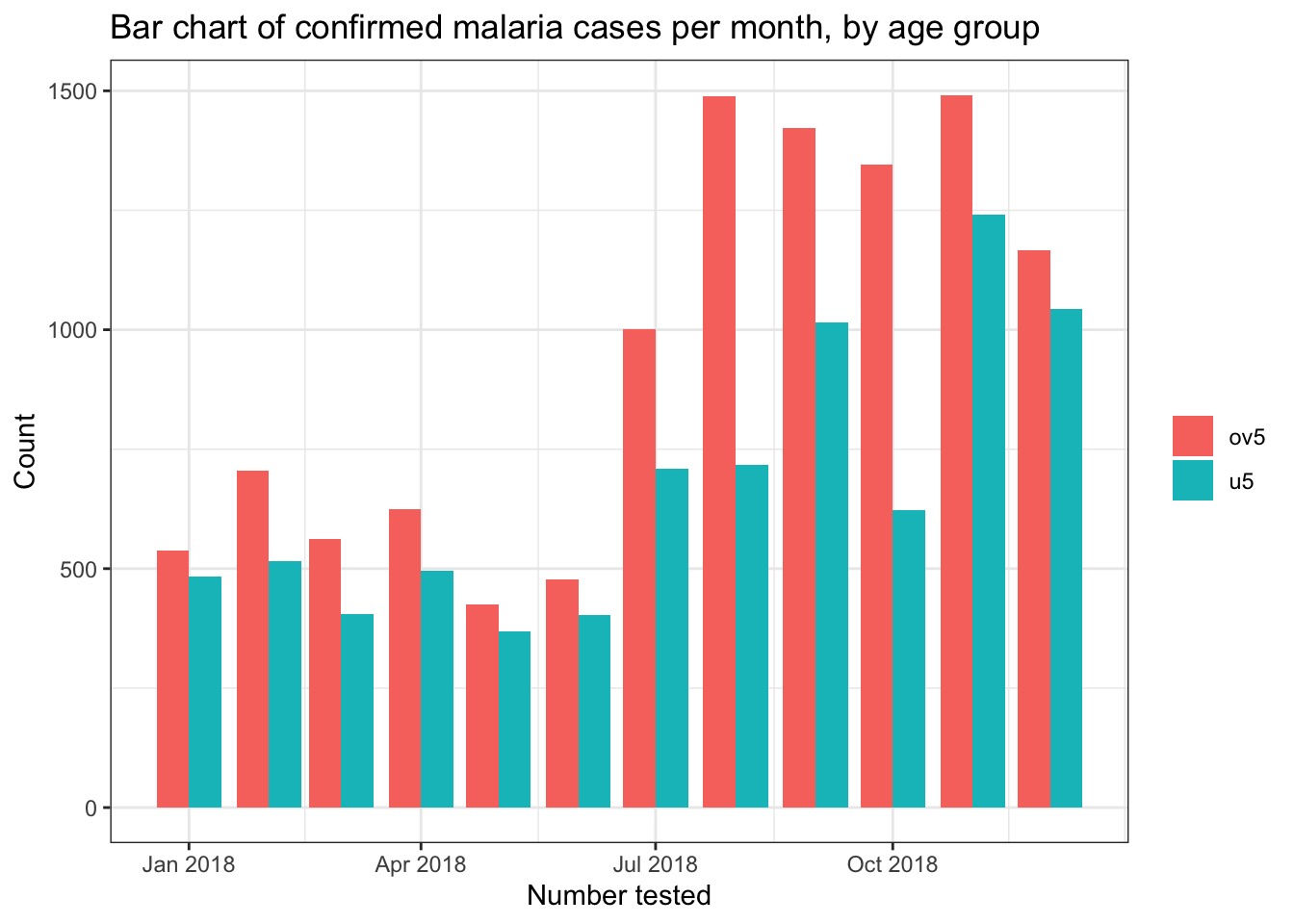
# Side by side bar chart
filter(pf_incidence, age_group != 'total') %>%
ggplot(mapping = aes(x = date_tested, y = conf, fill = age_group))+
geom_bar(stat = "identity", position = "dodge")+
theme_bw()+
labs(x = "Number tested",
y = "Count",
title = "Bar chart of confirmed malaria cases per month, by age group",
fill = NULL) 
This is just an introduction to what you can achieve for ggplot. For further guidance look at the help pages and the cheat-sheet available at https://www.rstudio.com/resources/cheatsheets.
Saving plots
There are a couple of different ways you can save plots in R. Firstly
you can save them by opening a png(), jpeg()
or pdf() depending on the file type you want to save. These
commands contain the output file path and the desired height and width
of the figure (optional). It is then followed by the figure, and closing
down the file with dev.off().
png("outputs/malaria_incidence_plot.png")
filter(pf_incidence, age_group != 'total') %>%
ggplot(mapping = aes(x = date_tested, y = conf, fill = age_group))+
geom_bar(stat = "identity", position = "dodge")+
theme_bw()+
labs(x = "Number tested",
y = "Count",
title = "Bar chart of confirmed malaria cases per month, by age group",
fill = NULL)
dev.off()Another option for saving plots is using ggsave(). This
takes the file name, including the extension, that you wish to save and
the plot. Leaving the plot name empty will default to the last plot
created. You can also include commands for the desired height and width
of the figure if required. So saving the above plot would entail:
filter(pf_incidence, age_group != 'total') %>%
ggplot(mapping = aes(x = date_tested, y = conf, fill = age_group))+
geom_bar(stat = "identity", position = "dodge")+
theme_bw()+
labs(x = "Number tested",
y = "Count",
title = "Bar chart of confirmed malaria cases per month, by age group",
fill = NULL)
ggsave("outputs/malaria_incidence_plot.png")Task 8
- Import the
pf_incidencedataset we created in demo 2.- Create a box plot of the confirmed malaria cases for children under 5 by month
- Using the
pf_incidencedataset create a stacked bar chart of confirmed > malaria cases in children under 5 and over 5’s by monthSolution
pf_incidence <- read_csv("outputs/pf_incidence.csv") %>% mutate(month = month(date_tested, label = TRUE, abbr = TRUE), year = year(date_tested)) pf_incidence %>% filter(age_group == "u5") %>% ggplot()+ geom_boxplot(mapping = aes(x = month, y = conf))+ labs(y = "Confirmed Cases")+ theme_bw() ggplot(pf_incidence)+ geom_boxplot(mapping = aes(x = month, y = conf, fill = age_group))+ labs(y = "Confirmed Cases under 5")+ theme_bw()