
Introduction to Spatial Analysis for Infectious Diseases
Welcome to the Introduction of Spatial Analysis for Infectious diseases tutorial pages. In this part of the workshop we aim to teach basic concepts, skills and tools for working with data so that you can get more done in less time, and with less pain. The lessons here focus mainly on the use of R and QGIS.
The main sections of this workshop are designed to be introductory
for participants with little to no programming experience. The advanced
section taught in R for Geostatistical Modelling using
R-INLA requires participants to have some statistical
knowledge, some understanding of Bayesian statistics and some prior
knowledge of spatial data.
Goal of this tutorial:
We want to provide our audience with a few useful tools for their day-to-day data activities. Given the restricted time of ASTMH, we may not be able to cover all important aspects of data handling. But the team are excited to release future workshops more tailored to regional/country requests. Please get in touch with us at on our GoogleGroup
Download the data
All data for this workshop will be available on a USB stick provided in attendance. The data can also be found in advance of the training in a GoogleDrive folder.
About the data
Notes
We have set up a Googledoc where we will put all the R scripts and notes we make during the workshop
Preparations
This workshops is expected to be hands-on, and to follow this lessons participants must have R, RStudio and QGIS installed on their computers. You also need to be able to install a number of R packages, create directories and download files.
To avoid troubleshooting during the lesson, participants should follow the instructions below to download and install everything beforehand. If you are using your own computers this should be no problem, but if the computer is managed by your organization’s IT department you might need help from an IT administrator.
Installing QGIS
Follow the QGIS download for the appropriate operating system to download on your computer.
- Scroll down and click on “QGIS Standalone Installer Version 3.26” to download the installer package: this is recommended for new users
- Save the window installer package in you desired location, usually on the C drive (“C:/Program Files/QGIS 3.26.2/”)
- Double click the installer and follow the prompts to install QGIS
Install R and RStudio
R and RStudio are two separate pieces of software:
- R is a programming language that is especially powerful for data exploration, visualization, and statistical analysis
- RStudio is an integrated development environment (IDE) that makes using R easier. In this course we use RStudio to interact with R.
If you don’t already have R and RStudio installed, follow the instructions for your operating system below. You have to install R before you install RStudio.
Windows
- Download R from the CRAN website.
- Run the
.exefile that was just downloaded - Go to the RStudio download page
- Under All Installers, download the RStudio Installer for Windows.
- Double click the file to install it
- Once it’s installed, open RStudio to make sure it works and you don’t get any error messages.
MacOS
- Download R from the CRAN website.
- Select the
.pkgfile for the latest R version - Double click on the downloaded file to install R
- It is also a good idea to install XQuartz (needed by some packages)
- Go to the RStudio download page
- Under All Installers, download the RStudio Installer for MacOS.
- Double click the file to install RStudio
- Once it’s installed, open RStudio to make sure it works and you don’t get any error messages.
Linux
- Follow the instructions for your distribution from CRAN, they provide
information to get the most recent version of R for common
distributions. For most distributions, you could use your package
manager (e.g., for Debian/Ubuntu run
sudo apt-get install r-base, and for Fedorasudo yum install R), but we don’t recommend this approach as the versions provided by this are usually out of date. In any case, make sure you have at least R 3.3.1. - Go to the RStudio download page
- Under All Installers, select the version that matches your
distribution and install it with your preferred method (e.g., with
Debian/Ubuntu
sudo dpkg -i rstudio-YYYY.MM.X-ZZZ-amd64.debat the terminal). - Once it’s installed, open RStudio to make sure it works and you don’t get any error messages.
Update R and RStudio
If you already have R and RStudio installed, first check if your R version is up to date:
- When you open RStudio your R version will be printed in the console
on the bottom left. Alternatively, you can type
sessionInfo()into the console. If your R version is 4.0.0 or later, you don’t need to update R for this lesson. If your version of R is older than that, download and install the latest version of R from the R project website for Windows, for MacOS, or for Linux - It is not necessary to remove old versions of R from your system, but if you wish to do so you can check How do I uninstall R?
- Note: The changes introduced by new R versions are usually
backwards-compatible. That is, your old code should still work after
updating your R version. However, if breaking changes happen, it is
useful to know that you can have multiple versions of R installed in
parallel and that you can switch between them in RStudio by going to
Tools > Global Options > General > Basic. - After installing a new version of R, you will have to reinstall all
your packages with the new version. For Windows, there is a package
called
installrthat can help you with upgrading your R version and migrate your package library.
To update RStudio to the latest version, open RStudio and click on
Help > Check for Updates. If a new version is available
follow the instruction on screen. By default, RStudio will also
automatically notify you of new versions every once in a while.
Install required R packages
During the course we will need a number of R packages. Packages
contain useful R code written by other people. We will use the packages
tidyverse, sf and terra mainly
for this workshop.
To try to install these packages, open RStudio. If this is your first
time opening it you may be prompted to set a CRAN mirror - select any
country or even the R global cran. This step is important because CRAN
aka Comprehensive R Archive Network is where R stores many
useful packages that we may use. If you forgot to set this up or wish to
change yours you can go to Tools ->
Global Options -> Packages, you will have a
screen that looks like this:
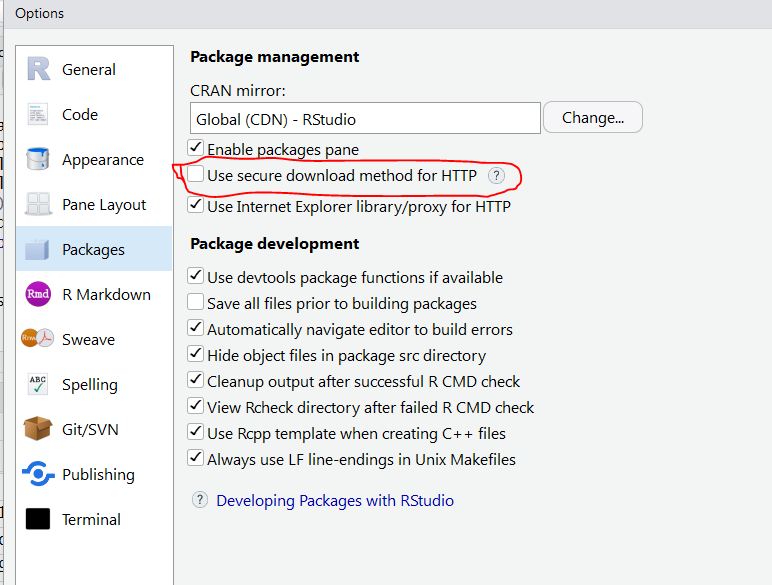
You can change CRAN by clicking the change... button.
Additionally, make sure that the section circled in red in the image
above is not selected
Next, copy and paste the following command into the console window (look for a blinking cursor on the bottom left), then press the Enter (Windows and Linux) or Return (MacOS) to execute the command.
list.of.packages <- c("tidyverse", "sf", "terra", "rgdal", "sp", "raster", "scico", "RColorBrewer", "viridis", "viridisLite", "ggplot2", "tidyr", "dplyr", "readxl")
new.packages <- list.of.packages[!(list.of.packages %in% installed.packages()[,"Package"])]
if(length(new.packages)) install.packages(new.packages)Alternatively, you can install the packages using RStudio’s graphical
user interface by going to Tools > Install Packages and
typing the names of the packages separated by a comma.
R tries to download and install the packages on your machine. Tidyverse is a group of packages so it can take some time to load. When the installation has finished, you can try to load the packages by pasting the following code into the console:
library(tidyverse)
library(sf)
library(terra)
library(rgdal)If you do not see an error like
there is no package called ‘...’ you are good to go!
Also note that when you load a package like tidyverse in for the first time you may see such test in your console:
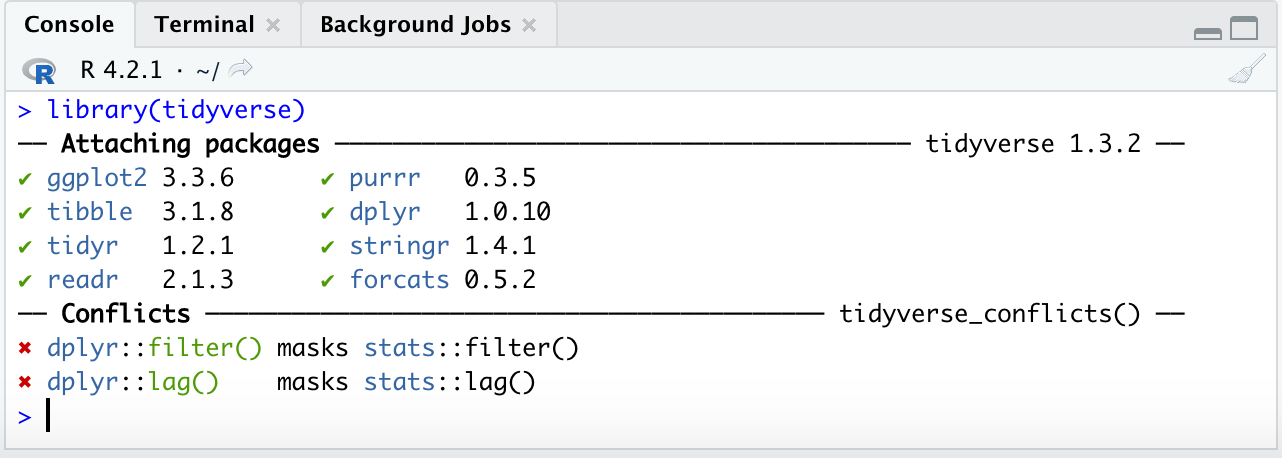
This text is simply telling you what packages have loaded under tidyverse and also telling you that there were function in base R that had the same names as those in tidyverse and they have been ‘over-written’ by the tidyverse ones. Additionally some of you may see some warning signs:
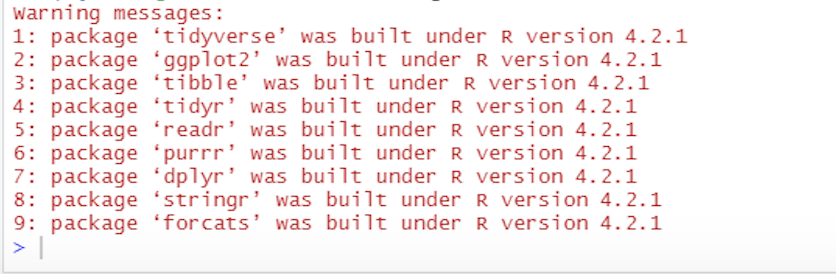
Warnings are normally ok to ignore. Usually when you see
a warning in R, it will still process the command you’ve entered, and
just wants to draw your attention that later on you may have issues
because of the text in the warning. Please ignore any warnings you may
see.
Updating R packages
Generally, it is recommended to keep your R version and all packages
up to date, because new versions bring improvements and important
bugfixes. To update the packages that you have installed, click
Update in the Packages tab in the bottom right
panel of RStudio, or go to
Tools > Check for Package Updates....
Sometimes, package updates introduce changes that break your old
code, which can be very frustrating. To avoid this problem, you can use
a package called renv. It locks the package versions you
have used for a given project and makes it straightforward to reinstall
those exact package version in a new environment, for example after
updating your R version or on another computer. However, the details are
outside of the scope of this lesson.